Abstract
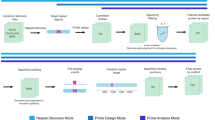
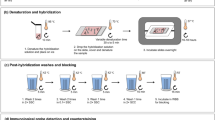
During the last decade, genome sequence databases of many species have been more and more completed so that it has become possible to further develop a recently established technique of FISH (Fluorescence In Situ Hybridization) called COMBO-FISH (COMBinatorial Oligo FISH). In contrast to standard FISH techniques, COMBO-FISH makes use of a bioinformatic search in sequence databases for probe design, so that it can be done for any species so far sequenced. In the original approach, oligonucleotide stretches of typical lengths of 15–30 nucleotides were selected in such a way that they only co-localize at the given genome target. Typical probe sets of about 20–40 stretches were used to label about 50–250 kb specifically. The probes of different lengths can be composed of purines and pyrimidines, but were often restricted to homo-purine or homo-pyrimidine probe sets because of the experimental advantage of using a protocol omitting denaturation of the target strand and triple strand binding of the probes. This allows for a better conservation of the 3D folding and arrangement of the genome. With an improved, rigorous genome sequence database analysis and sequence search according to statistical frequency and uniqueness, a novel family of probes repetitively binding to characteristic genome features like SINEs (Short Interspersed Nuclear Elements, e.g., ALU elements), LINEs (Long Interspersed Nuclear Elements, e.g., L1), or centromeres has been developed. These probes can be synthesized commercially as DNA or PNA probes with high purity and labeled by fluorescent dye molecules. Here, new protocols are described for purine-pyrimidine probes omitting heat treatment for denaturation of the target so that oligonucleotide labeling can also be combined with immune-staining by specific antibodies. If the dyes linked to the oligonucleotide stretches undergo reversible photo-bleaching (laser-induced slow blinking), the labeled cell nuclei can be further subjected to super-resolution localization microscopy for complex chromatin architecture research.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Bridger JM, Volpi EV (eds) (2010) Fluorescence in situ hybridization (FISH) – protocols and applications. Humana Press, Springer, New York
Wolf D, Rauch J, Hausmann M, Cremer C (1999) Comparison of the thermal denaturation behaviour of DNA-solutions and chromosome preparations in suspension. Biophys Chem 81:207–221
Rauch J, Wolf D, Hausmann M, Cremer C (2000) The influence of formamide on thermal denaturation profiles of DNA and metaphase chromosomes in suspensions. Z Naturforsch C 55:737–746
Winkler R, Perner B, Rapp A, Durm M, Cremer C, Greulich KO, Hausmann M (2003) Labelling quality and chromosome morphology after low temperature FISH analysed by scanning far-field and scanning near-field optical microscopy. J Microsc 209:23–33
Hausmann M, Winkler R, Hildenbrand G, Finsterle J, Weisel A, Rapp A, Schmitt E, Janz S, Cremer C (2003) COMBO-FISH: specific labelling of nondenatured chromatin targets by computer-selected DNA oligonucleotide probe combinations. Biotechniques 35:564–577
Schmitt E, Schwarz-Finsterle J, Stein S, Boxler C, Müller P, Mokhir A, Krämer R, Cremer C, Hausmann M (2010) Combinatorial Oligo FISH: directed labelling of specific genome domains in differentially fixed cell material and live cells. In: Bridger JM, Volpi EV (eds) Fluorescence in situ hybridization (FISH) – protocols and applications, Methods in molecular biology, vol 659. Humana Press, New York, pp 185–202
Zeller D, Kepper N, Hausmann M, Schmitt E (2013) Sequential and structural biophysical aspects of combinatorial oligo-FISH in Her2/neu breast cancer diagnostics. IFMBE Proc 38:82–85
Schwarz-Finsterle J, Stein S, Großmann C, Schmitt E, Trakhtenbrot L, Rechavi G, Amariglio N, Cremer C, Hausmann M (2007) Comparison of triplehelical COMBO-FISH and standard FISH by means of quantitative microscopic image analysis of abl/bcr genome organisation. J Biophys Biochem Methods 70:397–406
Schwarz-Finsterle J, Stein S, Großmann C, Schmitt E, Schneider H, Trakhtenbrot L, Rechavi G, Amariglio N, Cremer C, Hausmann M (2005) COMBO-FISH for focussed fluorescence labelling of gene domains: 3D-analysis of the genome architecture of abl and bcr in human blood cells. Cell Biol Int 29:1038–1046
Müller P, Rößler J, Schwarz-Finsterle J, Pedersen EB, Géci I, Schmitt E, Hausmann M (2016) PNA-COMBO-FISH: from combinatorial probe design in silico to vitality compatible, specific labelling of gene targets in cell nuclei. Exp Cell Res 345:51–59
Nolte O, Müller M, Häfner B, Knemeyer J-P, Stöhr K, Wolfrum J, Hakenbeck R, Denapaite D, Schwarz-Finsterle J, Stein S, Schmitt E, Cremer C, Herten D-P, Hausmann M, Sauer M (2006) Novel singly labelled probes for identification of microorganisms, detection of antibiotic resistance genes and mutations, and tumor diagnosis (SMART PROBES). In: Popp J, Strehle M (eds) Biophotonics. Wiley-VCH, Weinheim, pp 167–230
Grossmann C, Schwarz-Finsterle J, Schmitt E, Birk U, Hildenbrand G, Cremer C, Trakhtenbrot L, Hausmann M (2010) Variations of the spatial fluorescence distribution in ABL gene chromatin domains measured in blood cell nuclei by SMI microscopy after COMBO – FISH labelling. In: Méndez-Vilas A, Díaz Álvarez J (eds) Microscopy: science, technology, applications and education, vol 1. Formatex Research Center, Badajoz, pp 688–695
Lee J-H, Laure Djikimi Tchetgna F, Krufczik M, Schmitt E, Cremer C, Bestvater F, Hausmann M (2019) COMBO-FISH: a versatile tool beyond standard FISH to study chromatin organization by fluorescence light microscopy. OBM Genet 3(1). https://doi.org/10.21926/obm.genet.1901064
Cremer C, Kaufmann R, Gunkel M, Pres S, Weiland Y, Müller P, Ruckelshausen T, Lemmer P, Geiger F, Degenhard M, Wege C, Lemmermann N, Holtappels R, Strickfaden H, Hausmann M (2011) Superresolution imaging of biological nanostructures by Spectral Precision Distance Microscopy (SPDM). Review. Biotechnol J 6:1037–1051
Lemmer P, Gunkel M, Baddeley D, Kaufmann R, Urich A, Weiland Y, Reymann J, Müller P, Hausmann M, Cremer C (2008) SPDM – light microscopy with single molecule resolution at the nanoscale. Appl Phys B 93:1–12
Lemmer P, Gunkel M, Weiland Y, Müller P, Baddeley D, Kaufmann R, Urich A, Eipel H, Amberger R, Hausmann M, Cremer C (2009) Using conventional fluorescent markers for far-field fluorescence localization nanoscopy allows resolution in the 10 nm range. J Microsc 235:163–171
Müller P, Schmitt E, Jacob A, Hoheisel J, Kaufmann R, Cremer C, Hausmann M (2010) COMBO-FISH enables high precision localization microscopy as a prerequisite for nanostructure analysis of genome loci. Int J Mol Sci 11:4094–4105
Stuhlmüller M, Schwarz-Finsterle J, Fey E, Lux J, Bach M, Cremer C, Hinderhofer K, Hausmann M, Hildenbrand G (2015) In situ optical sequencing and nano-structure analysis of a trinucleotide expansion region by localization microscopy after specific COMBO-FISH labelling. Nanoscale 7:17938–17946. https://doi.org/10.1039/C5NR04141D
Schmitt E, Wagner J, Hausmann M (2012) Combinatorial selection of short triplex forming oligonucleotides for fluorescence in situ hybridisation COMBO-FISH. J Comput Sci 3:328–334. https://doi.org/10.1016/j.jocs.2011.10.001
Kepper N, Schmitt E, Lesnussa M, Weiland Y, Eussen HB, Grosveld F, Hausmann M, Knoch TA (2010) Visualization, analysis, and design of COMBO-FISH probes in the Grid-based GLOBE 3D genome platform. In: Solomonides T, Blanquer I, Breton V, Glatard T, Legré Y (eds) Healthgrid applications and core technologies. Proceedings of HealthGrid 2010, studies in health technology and informatics, vol 159, pp 171–180. ISBN: 978-1-60750-582-2
Sievers A, Bosiek K, Bisch M, Dreessen C, Riedel J, Froß P, Hausmann M, Hildenbrand G (2017) Kmer content, correlation and position analysis of genome DNA sequences for identification of function and evolutionary features. Genes 8:122. https://doi.org/10.3390/genes8040122
Sievers A, Wenz F, Hausmann M, Hildenbrand G (2018) Conservation of k-mer composition and correlation contribution between introns and intergenic regions of animalia genomes. Genes 9:482. https://doi.org/10.3390/genes9100482
Krufczik M, Sievers A, Hausmann A, Lee J-H, Hildenbrand G, Schaufler W, Hausmann M (2017) Combining low temperature fluorescence DNA-hybridization, immunostaining, and super-resolution localization microscopy for nano-structure analysis of ALU elements and their influence on chromatin structure. Int J Mol Sci 18:1005. https://doi.org/10.3390/ijms18051005
Hausmann M, Ilić N, Pilarczyk G, Lee J-H, Logeswaran A, Borroni AP, Krufczik M, Theda F, Waltrich N, Bestvater F, Hildenbrand G, Cremer C, Blank M (2017) Challenges for super-resolution localization microscopy and biomolecular fluorescent nano-probing in cancer research. Int J Mol Sci 18:2066. https://doi.org/10.3390/ijms18102066
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Hausmann, M., Lee, JH., Sievers, A., Krufczik, M., Hildenbrand, G. (2020). COMBinatorial Oligonucleotide FISH (COMBO-FISH) with Uniquely Binding Repetitive DNA Probes. In: Hancock, R. (eds) The Nucleus . Methods in Molecular Biology, vol 2175. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-0763-3_6
Download citation
DOI: https://doi.org/10.1007/978-1-0716-0763-3_6
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-0762-6
Online ISBN: 978-1-0716-0763-3
eBook Packages: Springer Protocols