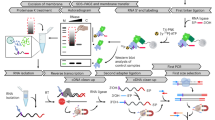
Abstract
In recent years, it has become increasingly recognized that regulation at the RNA level pervasively shapes the transcriptome in eukaryotic cells. This has fostered an interest in the mode of action of RNA-binding proteins that, via interaction with specific RNA sequence motifs, modulate gene expression. Understanding such posttranscriptional networks controlled by an RNA-binding protein requires a comprehensive identification of its in vivo targets. In metazoans and yeast, methods have been devised to stabilize RNA-protein interactions by UV cross-linking before isolating RNA-protein complexes using antibodies, followed by identification of associated RNAs by next-generation sequencing. These methods are collectively referred to as CLIP-Seq (cross-linking immunoprecipitation-high-throughput sequencing). Here, we present a version of the individual nucleotide resolution cross-linking and immunoprecipitation procedure that is suitable for use in the model plant Arabidopsis thaliana.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Singh G, Pratt G, Yeo GW et al (2015) The clothes make the mRNA: past and present trends in mRNP fashion. Annu Rev Biochem 84:325–354
Staiger D (2001) RNA-binding proteins and circadian rhythms in Arabidopsis thaliana. Philos Trans R Soc Lond B Biol Sci 356:755–1759
Kupsch C, Ruwe H, Gusewski S et al (2012) Arabidopsis chloroplast RNA binding proteins CP31A and CP29A associate with large transcript pools and confer cold stress tolerance by influencing multiple chloroplast RNA processing steps. Plant Cell 24:4266–4280
Kang H, Park SJ, Kwak KJ (2013) Plant RNA chaperones in stress response. Trends Plant Sci 18:100–106
Lewinski M, Hallmann A, Staiger D (2016) Genome-wide identification and phylogenetic analysis of plant RNA binding proteins comprising both RNA recognition motifs and contiguous glycine residues. Mol Gen Genomics 291:763–773
Rataj K, Simpson GG (2014) Message ends: RNA 3′ processing and flowering time control. J Exp Bot 65:353–363
Mili S, Steitz JA (2004) Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA 10:1692–1694
Xing D, Wang Y, Hamilton M et al (2015) Transcriptome-wide identification of RNA targets of Arabidopsis SERINE/ARGININE-RICH45 uncovers the unexpected roles of this RNA binding protein in RNA processing. Plant Cell 27:3294–3308
Köster T, Meyer K (2018) Plant ribonomics: proteins in search of RNA partners. Trends Plant Sci 23:352–365
Köster T, Staiger D (2014) RNA-binding protein immunoprecipitation from whole-cell extracts. Methods Mol Biol 1062:679–695
Köster T, Haas M, Staiger D (2014) The RIPper case: identification of RNA-binding protein targets by RNA immunoprecipitation. Methods Mol Biol 1158:107–121
Meyer K, Köster T, Nolte C et al (2017) Adaptation of iCLIP to plants determines the binding landscape of the clock-regulated RNA-binding protein AtGRP7. Genome Biol 18:204
Zhang Y, Gu L, Hou Y et al (2015) Integrative genome-wide analysis reveals HLP1, a novel RNA-binding protein, regulates plant flowering by targeting alternative polyadenylation. Cell Res 25:864–876
Marondedze C, Thomas L, Serrano NL et al (2016) The RNA-binding protein repertoire of Arabidopsis thaliana. Sci Rep 6:29766
Reichel M, Liao Y, Rettel M et al (2016) In planta determination of the mRNA-binding proteome of Arabidopsis etiolated seedlings. Plant Cell 28:2435–2452
Zhang Z, Boonen K, Ferrari P et al (2016) UV crosslinked mRNA-binding proteins captured from leaf mesophyll protoplasts. Plant Methods 12:42
Köster T, Marondedze C, Meyer K et al (2017) RNA-binding proteins revisited – the emerging Arabidopsis mRNA interactome. Trends Plant Sci 22:512–526
Köster T, Reichel M, Staiger D (2019) CLIP and RNA interactome studies to unravel genome-wide RNA-protein interactions in vivo in Arabidopsis thaliana. Methods. https://doi.org/10.1016/j.ymeth.2019.09.005
Sugimoto Y, Konig J, Hussain S et al (2012) Analysis of CLIP and iCLIP methods for nucleotide-resolution studies of protein-RNA interactions. Genome Biol 13:R67
König J, Zarnack K, Rot G et al (2010) iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol 17:909–915
Wang Z, Kayikci M, Briese M et al (2010) iCLIP predicts the dual splicing effects of TIA-RNA interactions. PLoS Biol 8:e1000530
Zarnack K, König J, Tajnik M et al (2013) Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of alu elements. Cell 152:453–466
Huppertz I, Attig J, D’Ambrogio A et al (2014) iCLIP: Protein–RNA interactions at nucleotide resolution. Methods 65:274–287
Acknowledgments
We greatly appreciate valuable suggestions on the protocol by Drs. Julian König, Kathi Zarnack, and Michaela Müller Mc-Nicoll. This work was supported by the DFG (STA 653/6, STA653/9, and STA653/14).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Köster, T., Staiger, D. (2020). Plant Individual Nucleotide Resolution Cross-Linking and Immunoprecipitation to Characterize RNA-Protein Complexes. In: Heinlein, M. (eds) RNA Tagging. Methods in Molecular Biology, vol 2166. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-0712-1_15
Download citation
DOI: https://doi.org/10.1007/978-1-0716-0712-1_15
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-0711-4
Online ISBN: 978-1-0716-0712-1
eBook Packages: Springer Protocols