Abstract
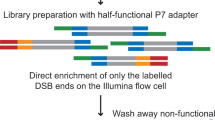
DNA double-strand breaks (DSBs) represent the most toxic form of DNA damage and can arise in either physiological or pathological conditions. If left unrepaired, these DSBs can lead to genome instability which serves as a major driver to tumorigenesis and other pathologies. Consequently, localizing DSBs and understanding the dynamics of break formation and the repair process are of great interest for dissecting underlying mechanisms and in the development of targeted therapies. Here, we describe END-seq, a highly sensitive next-generation sequencing technique for quantitatively mapping DNA double-strand breaks (DSB) at nucleotide resolution across the genome in an unbiased manner. END-seq is based on the direct ligation of a sequencing adapter to the ends of DSBs and provides information about DNA processing (end resection) at DSBs, a critical determinant in the selection of repair pathways. The absence of cell fixation and the use of agarose for embedding cells and exonucleases for blunting the ends of DSBs are key advances that contribute to the technique’s increased sensitivity and robustness over previously established methods. Overall, END-seq has provided a major technical advance for mapping DSBs and has also helped inform the biology of complex biological processes including genome organization, replication fork collapse and chromosome fragility, off-target identification of RAG recombinase and gene-editing nucleases, and DNA end resection at sites of DSBs.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Iacovoni JS et al (2010) High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J 29(8):1446–1457
Szilard RK et al (2010) Systematic identification of fragile sites via genome-wide location analysis of gamma-H2AX. Nat Struct Mol Biol 17(3):299–305
Yamane A et al (2011) Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nat Immunol 12(1):62–69
Barlow JH et al (2013) Identification of early replicating fragile sites that contribute to genome instability. Cell 152(3):620–632
Frock RL et al (2015) Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol 33(2):179–186
Tsai SQ et al (2015) GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol 33(2):187–197
Wang X et al (2015) Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nat Biotechnol 33(2):175–178
Crosetto N et al (2013) Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat Methods 10(4):361–365
Yan WX et al (2017) BLISS is a versatile and quantitative method for genome-wide profiling of DNA double-strand breaks. Nat Commun 8:15058
Longhese MP et al (2010) Mechanisms and regulation of DNA end resection. EMBO J 29(17):2864–2874
Canela A et al (2016) DNA breaks and end resection measured genome-wide by end sequencing. Mol Cell 63(5):898–911
Canela A et al (2017) Genome organization drives chromosome fragility. Cell 170(3):507–521. e18
Canela A et al (2019) Topoisomerase II-induced chromosome breakage and translocation is determined by chromosome architecture and transcriptional activity. Mol Cell 75(2):252–266. e8
Tubbs A et al (2018) Dual roles of poly(dA:dT) tracts in replication initiation and fork collapse. Cell 174(5):1127–1142. e19
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Wong, N., John, S., Nussenzweig, A., Canela, A. (2021). END-seq: An Unbiased, High-Resolution, and Genome-Wide Approach to Map DNA Double-Strand Breaks and Resection in Human Cells. In: Aguilera, A., Carreira, A. (eds) Homologous Recombination. Methods in Molecular Biology, vol 2153. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-0644-5_2
Download citation
DOI: https://doi.org/10.1007/978-1-0716-0644-5_2
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-0643-8
Online ISBN: 978-1-0716-0644-5
eBook Packages: Springer Protocols