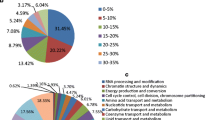
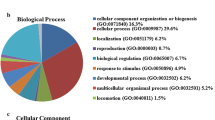
Abstract
Mass spectrometry-based proteomics analysis could categorize proteins and study their interactions in large scale in human cancers. By this method, many proteins are upregulated or downregulated in esophageal squamous cell carcinoma (ESCC) when compared to nonneoplastic esophageal mucosae. The method can also be used to identify novel, effective biomarkers for early diagnosis or predict prognosis of patients with ESCC. These changes are associated with different clinical and pathological parameters. Different biological matrices such as pathological tissue, body fluids, and cancer cell lines-based proteomics have widely been used. Herein, we described cell line-based label-free shotgun proteomics (in-solution tryptic digestion) to identify the protein biomarkers differently expressed in ESCC.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Anderson NL, Anderson NG (1998) Proteome and proteomics: new technologies, new concepts, and new words. Electrophoresis 19:1853–1861
Tyers M, Mann M (2003) From genomics to proteomics. Nature 422:193–197
Gronborg M, Kristiansen TZ, Iwahori A, Chang R, Reddy R, Sato N et al (2006) Biomarker discovery from pancreatic cancer secretome using a differential proteomic approach. Mol Cell Proteomics 5:157–171
Wu HY, Chang YH, Chang YC, Liao PC (2009) Proteomics analysis of nasopharyngeal carcinoma cell secretome using a hollow fiber culture system and mass spectrometry. J Proteome Res 8:380–389
Iannetti A, Pacifico F, Acquaviva R, Lavorgna A, Crescenzi E, Vascotto C et al (2008) The neutrophil gelatinase-associated lipocalin (NGAL), a NF-kappa-B-regulated gene, is a survival factor for thyroid neoplastic cells. Proc Natl Acad Sci U S A 105:14058–14063
Zhong L, Roybal J, Chaerkady R, Zhang W, Choi K, Alvarez CA et al (2008) Identification of secreted proteins that mediate cell-cell interactions in an in vitro model of the lung cancer microenvironment. Cancer Res 68:7237–7245
Weng LP, Wu CC, Hsu BL, Chi LM, Liang Y, Tseng CP et al (2008) Secretome-based identification of mac-2 binding protein as a potential oral cancer marker involved in cell growth and motility. J Proteome Res 7:3765–3775
Kobayashi R, Deavers M, Patenia R, Rice-Stitt T, Halbe J, Gallardo S et al (2009) 14-3-3 zeta protein secreted by tumor associated monocytes/macrophages from ascites of epithelial ovarian cancer patients. Cancer Immunol Immunother 58:247–258
Wu CC, Chen HC, Chen SJ, Liu HP, Hsieh YY, Yu CJ et al (2008) Identification of collapsin response mediator protein-2 as a potential marker of colorectal carcinoma by comparative analysis of cancer cell secretomes. Proteomics 8:316–332
Dombkowski AA, Cukovic D, Novak RF (2006) Secretome analysis of microarray data reveals extracellular events associated with proliferative potential in a cell line model of breast disease. Cancer Lett 241:49–58
Paulitschke V, Kunstfeld R, Mohr T, Slany A, Micksche M, Drach J et al (2009) Entering a new era of rational biomarker discovery for early detection of melanoma metastases: secretome analysis of associated stroma cells. J Proteome Res 8:2501–2510
Cravatt BF, Simon GM, Yates JR III (2007) The biological impact of mass-spectrometry-based proteomics. Nature 450:991–1000
Kashyap MK, Harsha HC, Renuse S, Pawar H, Sahasrabuddhe NA, Kim MS et al (2010) SILAC-based quantitative proteomic approach to identify potential biomarkers from the esophageal squamous cell carcinoma secretome. Cancer Biol Ther 10:796–810
Xue H, Lu B, Lai M (2008) The cancer secretome: a reservoir of biomarkers. J Transl Med 6:52
Zhang W, Matrisian LM, Holmbeck K, Vick CC, Rosenthal EL (2006) Fibroblast-derived MT1-MMP promotes tumor progression in vitro and in vivo. BMC Cancer 6:52
Du XL, Hu H, Lin DC, Xia SH, Shen XM, Zhang Y et al (2007) Proteomic profiling of proteins dysregulated in Chinese esophageal squamous cell carcinoma. J Mol Med 85:863–875
Fu L, Qin YR, Xie D, Chow HY, Ngai SM, Kwong DL et al (2007) Identification of alpha-actinin 4 and 67 kDa laminin receptor as stage-specific markers in esophageal cancer via proteomic approaches. Cancer 110:2672–2681
Fujita Y, Nakanishi T, Hiramatsu M, Mabuchi H, Miyamoto Y, Miyamoto A et al (2006) Proteomics-based approach identifying autoantibody against peroxiredoxin VI as a novel serum marker in esophageal squamous cell carcinoma. Clin Cancer Res 12:6415–6420
Fujita Y, Nakanishi T, Miyamoto Y, Hiramatsu M, Mabuchi H, Miyamoto A et al (2008) Proteomics-based identification of autoantibody against heat shock protein 70 as a diagnostic marker in esophageal squamous cell carcinoma. Cancer Lett 263:280–290
Uemura N, Nakanishi Y, Kato H, Saito S, Nagino M, Hirohashi S et al (2009) Transglutaminase 3 as a prognostic biomarker in esophageal cancer revealed by proteomics. Int J Cancer 124:2106–2115
Zhou G, Li H, Gong Y, Zhao Y, Cheng J, Lee P, Zhao Y (2005) Proteomic analysis of global alteration of protein expression in squamous cell carcinoma of the esophagus. Proteomics 5:3814–3821
Breton J, Gage MC, Hay AW, Keen JN, Wild CP, Donnellan C et al (2008) Proteomic screening of a cell line model of esophageal carcinogenesis identifies cathepsin D and aldo-keto reductase 1C2 and 1B10 dysregulation in Barrett’s esophagus and esophageal adenocarcinoma. J Proteome Res 7:1953–1962
Xu SY, Liu Z, Ma WJ, Sheyhidin I, Zheng ST, Lu XM (2009) New potential biomarkers in the diagnosis of esophageal squamous cell carcinoma. Biomarkers 14:340–346
Bauer KM, Lambert PA, Hummon AB (2012) Comparative label-free LC-MS/MS analysis of colorectal adenocarcinoma and metastatic cells treated with 5-fluorouracil. Proteomics 12:1928–1937
Islam F, Chaousis S, Wahab R, Gopalan V, Lam AK (2018) Protein interactions of FAM134B with EB1 and APC/beta-catenin in vitro in colon carcinoma. Mol Carcinog 57:1480–1491
Zhang LY, Ying WT, Mao YS, He HZ, Liu Y, Wang HX et al (2003) Loss of clusterin both in serum and tissue correlates with the tumorigenesis of esophageal squamous cell carcinoma via proteomics approaches. World J Gastroenterol 9:650–654
Qi Y, Chiu JF, Wang L, Kwong DL, He QY (2005) Comparative proteomic analysis of esophageal squamous cell carcinoma. Proteomics 5:2960–2971
Wang SJ, Zhang LW, Yu WF, Yu JK, Zheng S, Li YS et al (2007) Establishment of a diagnostic model of serum protein fingerprint pattern for esophageal cancer screening in high incidence area and its clinical value. Zhonghua Zhong Liu Za Zhi 29:441–443
Liu WL, Zhang G, Wang JY, Cao JY, Guo XZ, Xu LH et al (2008) Proteomics-based identification of autoantibody against CDC25B as a novel serum marker in esophageal squamous cell carcinoma. Biochem Biophys Res Commun 375:440–445
Shao CCS, Chen L, Cobos E, Wang J, Haab BB, Gao W (2009) Antibody microarray analysis of serum glycans in esophageal squamous cell carcinoma cases and controls. Proteomics Clin Appl 3:923–931
Chen JY, Xu L, Fang WM, Han JY, Wang K, Zhu KS (2017) Identification of PA28β as a potential novel biomarker in human esophageal squamous cell carcinoma. Tumour Biol 39:1010428317719780
Pawar H, Kashyap MK, Sahasrabuddhe NA, Renuse S, Harsha HC, Kumar P et al (2011) Quantitative tissue proteomics of esophageal squamous cell carcinoma for novel biomarker discovery. Cancer Biol Ther 12:510–522
Yazdian-Robati R, Ahmadi H, Riahi MM, Lari P, Aledavood SA, Rashedinia M et al (2017) Comparative proteome analysis of human esophageal cancer and adjacent normal tissues. Iran J Basic Med Sci 20:265–271
Zhao J, Fan YX, Yang Y, Liu DL, Wu K, Wen FB et al (2015) Identification of potential plasma biomarkers for esophageal squamous cell carcinoma by a proteomic method. Int J Clin Exp Pathol 8:1535–1544
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Islam, F., Gopalan, V., Lam, A.K. (2020). Mass Spectrometry for Biomarkers Discovery in Esophageal Squamous Cell Carcinoma. In: Lam, A. (eds) Esophageal Squamous Cell Carcinoma. Methods in Molecular Biology, vol 2129. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-0377-2_19
Download citation
DOI: https://doi.org/10.1007/978-1-0716-0377-2_19
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-0376-5
Online ISBN: 978-1-0716-0377-2
eBook Packages: Springer Protocols