Abstract
T, B, and NK lymphocytes are generated from pluripotent hematopoietic stem cells through a successive series of lineage restriction processes. Many regulatory components, such as transcription factors, cytokines/cytokine receptors, and signal transduction molecules orchestrate cell fate specification and determination. In particular, transcription factors play a key role in regulating lineage-associated gene programs. Recent findings suggest the involvement of epigenetic factors in the maintenance of cell fate. Here, we review the early developmental events during lymphocyte lineage determination, focusing on the transcriptional networks and epigenetic regulation. Finally, we also discuss the developmental relationship between acquired and innate lymphoid cells.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Natural Killer Cell
- Notch Signaling
- Lymphocyte Development
- Innate Lymphoid Cell
- Lymphoid Tissue Induce Cell
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
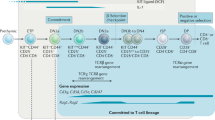
Lymphocytes consist of the T and B cells of the adaptive immune system as well as natural killer (NK) cells, lymphoid tissue inducing (LTi) cells, and natural helper (NH) cells, which have recently been defined as a new subset termed innate lymphoid cells (ILCs). All of these lymphocytes are derived from hematopoietic stem cells (HSCs) found in fetal liver before birth and in bone marrow (BM) after birth, although the developmental pathways and the molecular mechanisms of their diversification processes are still not completely understood. The lymphocytes are generated through a gain of specific gene expression signatures mediated by a combinatorial network of transcription factors. This is accompanied by the gradual loss of differentiation potential for alternative cell lineages (Yang et al. 2010; Mandel and Grosschedl 2010) (Fig. 1). Lineage-specific transcription factors play pivotal roles in the lineage restriction process. These transcription factors activate lineage-specific genes to promote lineage development, while simultaneously restricting the developmental potential of multipotent progenitors to the lymphoid pathway by suppressing the lineage-inappropriate genes, which are associated with other differentiation pathways. Recent findings also suggest that the downregulation of stem cell-associated genes by transcription factors is also important for lineage restriction programs (Nutt and Kee 2007; Rothenberg et al. 2008). Many transcription factors have been shown to be critical for the generation of lymphoid cells. However, how these transcription factors work together through regulatory networks and how the specific combinations of transcription factors within the networks synergize or antagonize each other are yet to be defined. In this review, we focus on the transcriptional networks and the epigenetic mechanisms that orchestrate lymphoid cell genesis and provide an overview of current models of the lymphoid lineage commitment process during immune cell development.
Schematic model of early lymphocyte development. Multipotent progenitors (purple) give rise to T cell (blue), B cell (green), and ILC (red) progenitors during specification to each lymphoid lineage. Transcription factors and epigenetic factors which are thought to be involved in cell fate specification and determination are indicated. Common innate lymphoid progenitors (ILCP) are still speculative, although Id2 is a master regulator of ILC development
2 Generation of T and B Cell Progenitors in BM
Many kinds of BM progenitors including HSCs and the downstream progenitors with T or B cell potential have been identified (Fig. 1). The earliest progenitors can be isolated based on the absence of lineage markers (Lin−) in combination with high expression of c-kit and Sca-1 (LSK cells) (Ikuta and Weissman 1992; Spangrude et al. 1988). This population can be further subdivided into at least three populations based on surface expression of CD34 (Osawa et al. 1996; Yamamoto et al. 2013) and fms-like tyrosine kinase receptor-3 (Flt-3) (Christensen and Weissman 2001; Adolfsson et al. 2001). CD34−Flt-3− LSK cells are defined as long-term repopulating HSCs that retain self-renewal activity and have the capacity to generate all blood lineages, including T and B cells . The immediate downstream progenitors of HSCs are the CD34+Flt-3− multipotent progenitors (MPPs) that have lost the ability to self-renew, but retain multilineage differentiation capability (Yang et al. 2005). CD34+Flt-3+ LSK cells are classified as lymphoid-primed MPPs (LMPPs), because they have lost megakaryocyte /erythroid potential and are biased toward lymphoid and granulocyte /macrophage lineages (Adolfsson et al. 2005). LMPPs contain lymphoid restricted cells, early lymphoid progenitors (ELPs), defined using reporter mice by expression of Rag1-GFP (Igarashi et al. 2002; Schwarz et al. 2007). Further downstream progenitors that retain T and B cell potential are termed common lymphoid progenitors (CLPs) (Kondo et al. 1997). CLPs were originally thought to be exclusively committed to T and B lineages, but are now suggested to be a rather heterogeneous population in terms of their differentiation potential. The Ly6D surface marker can separate the CLP population into two populations, the Ly6D− progenitors with B cell , T cell , NK cell, and dendritic cell (DC) potential and the Ly6D+ progenitors, which are largely restricted to the B cell lineage (Inlay et al. 2009). Fate mapping analysis also suggests that CLPs with a history of λ5 and Rag-1 expression are largely restricted to the B cell lineage (Mansson et al. 2010). Furthermore, as described below, the earliest lymphoid progenitors in the thymus retain limited B cell potential compared to the T or myeloid lineages (Wada et al. 2008; Kawamoto et al. 2010). Taken together, these results suggest that the CLP may not be the main source of thymic immigrants that contribute to the generation of T cells .
The first identifiable B cell -specific progenitors in BM are the Pre-Pro B cells or fraction A, which have the cell surface phenotype IgM−B220+CD43+CD19−HSA−BP1− (Hardy et al. 1991; Li et al. 1996). Pre-Pro B cells are specified but not totally committed to the B cell lineage. Cells committed to this lineage are called Pro B cells, which are identified by the expression of B220, CD43, and CD19.
3 Transcriptional Regulation During T Cell Lineage Commitment
3.1 T and Myeloid Potential are Closely Associated in Early Thymic Progenitors
Thymocytes can be separated into four populations based on the expression of CD4 and CD8. The CD4−CD8− double negative (DN) stage is the most immature stage among these four populations. DN cells develop into CD4+CD8+ double positive (DP) cells and finally mature into CD4+CD8− single positive (CD4SP) or CD4−CD8+ (CD8SP) cells. DN cells can be further subdivided into four populations based on the surface expression of CD44 and CD25: CD44+CD25+ (DN1), CD44+CD25+ (DN2), CD44−CD25+ (DN3), and CD44−CD25− (DN4) (Godfrey et al. 1993), and differentiation progresses in this order (Fig. 2). Previous work demonstrated that the c-kit+ cells among DN1 cells, also called early thymic progenitors (ETPs), were the most immature progenitors in the thymus (Matsuzaki et al. 1993; Allman et al. 2003; Wada et al. 2008). ETPs are not committed to the T cell lineage but maintain the potential to generate T cells , NK cells, myeloid cells, DCs, and B cells . However, we and other groups demonstrated that most of the earliest T cell progenitors in the DN1 subset have already lost B cell potential (Wada et al. 2008; Bell and Bhandoola 2008; Lu et al. 2005), moreover, the overall frequency of cells with B cell potential is much lower than that of cells with T or myeloid potential. To simultaneously assess myeloid and T cell potential, we developed a new culture system using TSt-4/DLL1 stromal cells, which ectopically express a Notch ligand DLL1. By culturing individual DN1 cells, we found that a substantial proportion of these thymic T cell progenitors retain myeloid potential. This observation was confirmed by other groups (Bell and Bhandoola 2008; Porritt et al. 2004; Balciunaite et al. 2005), who also reported that the T and myeloid lineages, but not the B cell lineage, are closely related to each other in the earliest T cell progenitors in adult thymus. Myeloid potential in the T cell progenitors was challenged by studies using fate mapping analysis with an IL7Rα-Cre driven reporter mice (Schlenner et al. 2010). These authors suggested a distinct origin of T and myeloid lineages in vivo based on the observation of a lower frequency of labeled myeloid cells than T cells in the thymus. However, 10–20 % of the macrophages and neutrophils in the thymus were marked by the history of IL-7R expression in this assay system. This observation is consistent with the previous report using a different in vivo system demonstrating that approximately 30 % of the macrophages are derived from early thymic T lineage progenitors in the steady state (Wada et al. 2008). Taken together, these results demonstrated that the T cell progenitors retain myeloid potential after terminating B cell potential, indicating that the CLPs are not the intermediate progenitors during T cell development.
3.2 Termination of Myeloid Potential in T Cell Progenitors
Since the earliest T cell progenitors in the thymus retain substantial myeloid potential, we wished to know when the myeloid potential is terminated. Using plckGFP Tg mice, in which GFP expression is regulated by the lck promoter, we have previously shown that DN2 cells can be separated into GFP− and GFP+ population (Masuda et al. 2007). GFP−DN2 cells retain myeloid cell, DC, and NK cell potential, while GFP+DN2 cells are restricted to the T lineage. Consistent with this observation, the expression of T lineage-associated genes, GATA3, CD3ε, and pre-Tα was upregulated in cells at the GFP+DN2 stage and, conversely, the myeloid transcription factor PU.1 was downregulated in these cells. We named GFP−DN2 cells as DN2mt (myeloid-T) and GFP+DN2 cells as DN2t (T lineage-committed) cells. Rothenberg et al. proposed a similar separation of DN2 cells based on the expression level of c-kit into ckithighCD25+DN2a and ckitlowCD25+DN2b cells (Rothenberg et al. 2008). Global gene expression analysis comparing these two populations suggests that DN2a precedes DN2b. These results indicate that the non-T lineage potential, myeloid and NK potential, of T cell progenitors is lost at the DN2 stage (Fig. 2).
To investigate the mechanism underlying termination of myeloid and NK potential, we have developed a novel T cell differentiation culture system (Ikawa et al. 2010). When murine hematopoietic progenitors were cultured on immobilized Notch ligand DLL4 in the presence of cytokines including IL-7, progenitors became arrested at the DN2mt stage. The arrested cells maintained an enormous proliferation capacity and differentiation potential to generate T cells as well as NK and myeloid lineage cells. Interestingly, withdrawal of IL-7 allowed the cells to commit to the T lineage, which could be recognized by the expression of plckGFP. Moreover, the committed T cell progenitors differentiated into DP cells under the same culture conditions. These results indicate that T cell differentiation can be induced in a stromal-free culture system by simply reducing the concentration of IL-7 in the middle of the culture period. Similar developmental arrest was observed in the thymus of mice lacking the transcription factor Bcl11b. The DN2 cells in Bcl11b-deficient mice can be propagated on TSt-4/DLL1 cells, retaining the potential to generate myeloid and NK lineage cells even after 1 month. Overexpression of Bcl11b in these cultured DN2 cells promoted the generation of DN3 cells, indicating the Bcl11b plays pivotal roles in T lineage determination (Fig. 2). Critical roles of Bcl11b in this T lineage checkpoint have also been demonstrated by other groups (Li et al. 2010a, b).
3.3 Transcriptional Regulation that Determines T Cell Fate
Many transcription factors including Bcl11b have been shown to be important for the T cell fate choice during the lineage determination process (Yang et al. 2010; Rothenberg 2012). These transcription factors function at different stages of T cell development and play different roles during T lineage commitment by coordinating the gene expression program (Fig. 3).
Transcriptional network during T cell lineage commitment. Notch signals from the thymic environment initiate the T lineage program. E2A/HEB act in synergy with Notch to promote T cell commitment. TCF-1 activates Bcl11b, which in turn suppresses the other lineage programs, such as myeloid and NK lineages
T cell factor 1 (TCF-1, encoded by the transcription factor 7 gene, Tcf7) is an essential transcription factor in early T cell differentiation (Verbeek et al. 1995; Hattori et al. 1996; Schilham et al. 1998). TCF-1 and its partner LEF-1 act as a transducer of Wnt pathway signals (Staal et al. 2008; Staal and Sen 2008). TCF1-deficient mice were originally shown to have a reduced number of thymocytes and a partial block of T cell development at the immature single positive (ISP) to DP stage (Schilham et al. 1998). However, recent findings suggest that TCF-1 is essential at the earliest stage of T cell specification in the thymus (Weber et al. 2011; Yu et al. 2012). These studies show that the expression of Tcf7 is directly activated by Notch signals and that TCF-1 in turn activates T lineage gene program including Bcl11b and GATA3. Forced expression of TCF-1 can bypass most of the requirement for Notch signaling and promote T lineage specification even without Notch signals in vitro. Interestingly, TCF-1 is also shown to be required for group 2 innate lymphoid cell (ILC2) development through GATA3-dependent and -independent pathways (Yang et al. 2013). ILC2 cells are innate lymphocytes that produce T helper type 2 (Th2) cell-associated cytokines interleukin (IL)-5 and IL-13 and mediate airway inflammation and helminth infection. TCF1-deficient mice lack ILC2 in the lung and BM. By using retroviral dominant-negative Mastermind (dnMAML), a pan Notch inhibitor, these investigators confirmed that the Notch signaling is required for ILC2 generation. Transduction of Tcf7 retrovirus partially restored the generation of ILC2 from dnMAML–expressing BM progenitors, indicating the TCF-1 acts downstream of Notch signaling during ILC2 development, similar to early T cell development. Thus, transcriptional regulatory elements that underlie early T cell development also induce the generation of ILC2, suggesting a close developmental relationship between T cells and innate lymphoid cells.
GATA3 is a zinc finger transcription factor that has also long been recognized as an important factor for the development of T cells at multiple checkpoints (Ting et al. 1996). GATA3 is indispensable for T cell development during the DN3 stage and for the generation of CD4SP cells (Pai et al. 2003). In addition, GATA3 is a master regulator of Th2 cell function (Zheng and Flavell 1997; Zhu et al. 2004). Several findings have demonstrated the critical importance of GATA3 for early T cell development (Hattori et al. 1996; Taghon et al. 2007). The expression of GATA3 initiates at the DN1 stage and is sustained throughout T cell development. Hosoya et al. have recently shown that GATA3 plays an essential role in DN1 cell generation (Hosoya et al. 2009). By contrast, normal numbers of prethymic progenitors, LMPPs and CLPs, were observed in GATA3-deficient fetal livers, indicating the critical role of GATA3 for the development of early thymic T cell progenitors.
E proteins are helix-loop-helix transcription factors that are essential for T and B lymphocyte development (de Pooter and Kee 2010). There are four E proteins in mammals, E2A (E12 and E47, encoded by the Tcf3 gene), HEB (Tcf12), and E2-2 (Tcf4) (Massari and Murre 2000). They function as homodimers or heterodimers in regulating differentiation at various stages of lymphocyte development. E2A/HEB heterodimers especially are required for early T cell development. Loss of E2A or HEB results in a partial block of early T cell development at the DN1 or ISP stages. Deletion of both E2A and HEB leads to a severe block at the DN stage, prior to pre-TCR expression, indicating the cooperation of these proteins for normal T cell development in the thymus. Association of E2A with other critical factors has also been shown. E2A is required to limit GATA3 expression specifically at the DN2 stage (Xu et al. 2013), thus elevated expression of GATA3 is seen in E2A-deficient DN2 cells. Ectopic GATA3 siRNA expression restores the DN3-like cell development by E2A-deficient MPPs, indicating that suppression of GATA3 by E2A is required for proper T cell development. E2A has also been shown to cooperate with Notch signaling to promote early T lineage commitment (Ikawa et al. 2006). Expression of several Notch-related genes, Notch1, Notch3, Hes1, and Deltex1 are directly regulated by E2A and once the Notch genes are activated, E2A and Notch act in synergy to promote T cell development. E2A continuously regulates the expression of Notch1 until the DN3 stage (Yashiro-Ohtani et al. 2009). E2A also plays critical roles in T cell receptor (TCR) β gene rearrangement and allelic exclusion (Agata et al. 2007). In addition to its essential role in intrathymic development, E2A is also known to be important for normal development of prethymic progenitors. LMPPs, which can be identified in the LSK compartment by surface expression of Flt3, are severely reduced in the absence of E2A (Dias et al. 2008; Semerad et al. 2009). E2A-deficient LMPPs show reduced expression of lymphoid-related genes, Notch1, Rag1, and CCR9. The combined activities of E2A and HEB are required for the induction of forkhead box O1 (FOXO1) expression at the CLP stage (Welinder et al. 2011). Together, these results suggest that E proteins play critical roles in both prethymic and intrathymic T cell development by inducing different combinatorial targets.
4 Transcriptional Regulation of B Cell Lineage Commitment
The process of commitment to the B cell lineage is also dependent on a network of transcription factors (Nutt and Kee 2007; Mandel and Grosschedl 2010). Gene targeting studies have demonstrated that several transcription factors, including Ikaros, PU.1, LRF, Bcl11a, Runx1, E2A, EBF1, and Pax5 play critical roles in inducing B lineage specification and determination, although most of the transcription factors except EBF1 and Pax5 are also essential for the development of hematopoietic stem/progenitors, T cells or myeloid cells. Moreover, while it was initially thought that these factors play different functions at different developmental stages, it has recently become clearer that these transcription factors act in a complex network to regulate the cell fate choices in a synergistic or antagonistic manner.
PU.1 is one of the key players in the gene regulatory networks controlling cell fate determination of lymphoid versus myeloid lineages, probably at the LMPP stage (de Koter and Singh 2000; Kueh et al. 2013). It has been suggested that the cell fate choice critically depends on PU.1 levels. The downregulation of PU.1 is required for B cell development, whereas higher levels favor the development of myeloid lineage cells. Recent findings suggest that the cell cycle status plays a critical role for inducing positive PU.1 feedback loops, which may lead to the myeloid fate choice (Kueh et al. 2013).
LRF (leukemia/lymphoma-related factor) has been suggested to act upstream of E2A and EBF1. LRF is a POZ and Kruppel (POK)-type transcription factor with multiple functions in hematopoietic/immune systems (Lunardi et al. 2013). Notably, inactivation of LRF promotes T cell fate at the expense of B cell development, suggesting that LRF regulates the Notch-mediated lineage decision (Maeda et al. 2007). However, a recent study demonstrated that the promotion of T lineage cells in BM of LRF-deficient mice was due to the aberrant expression of DLL4 in erythroblasts, suggesting that LRF does not directly regulate T or B lineage fate choices under physiological conditions (Lee et al. 2013).
Bcl11a also functions upstream of E2A and EBF1 and is known to regulate lymphoid and erythroid development (Durum 2003). Loss of Bcl11a results in a lack of B cells and impaired thymocyte maturation (Liu et al. 2003). Recent findings also demonstrated a critical role of Bcl11a for pDC development by regulating Flt3 expression. The numbers of LSK cells and CLPs were greatly reduced in both fetal liver and BM of Bcl11a-deficient mice. In addition, expression of Flt3 and IL-7R was reduced in MPPs in BM of Bcl11a-deficient mice, suggesting a critical role of Bcl11a in the development and maintenance of LMPPs and CLPs (Wu et al. 2013). This observation is intriguing in that Bcl11a shares many features with another Kruppel-like transcription factor, Bcl11b, which is critical for T lineage commitment as described above. Bcl11a and Bcl11b may function in an antagonistic manner to regulate T or B lineage development. It would also be interesting to clarify the relationship between Bcl11a and other transcription factors, Ikaros and PU.1, which play critical roles at similar developmental stages.
As noted above, E proteins (E2A, HEB, and E2-2) are required for maintaining HSC pools and promoting the development of lymphoid progenitors (Dias et al. 2008; Semerad et al. 2009). The E2A proteins act upstream and in concert with EBF1 and Pax5 to promote specification and determination to the B cell lineage. E2A and HEB cooperatively induce the expression of FOXO1 at the CLP stage (Welinder et al. 2011). A requirement of E2A for commitment to the B lineage was also shown by establishment of long-term cultured multipotent progenitors derived from E2A-deficient BM (Ikawa et al. 2004). E2A-deficient hematopoietic progenitors (HPCs) can efficiently grow on S17 stromal cells in the presence of SCF, IL-7, and Flt3-Ligand, and the cells maintain the potential to generate T cells , NK cells, myeloid cells, DCs, and erythroid cells. Enforced expression of E47 in the E2A-deficient HPCs directly activates the transcription of a subset of B cell -specific genes, including λ5, mb-1, and Pax5. By contrast, E47 inhibits the expression of genes involved in the differentiation of other lineages, TCF-1 and GATA1. These results indicate that E2A plays a critical role in B lineage commitment.
Once E2A activates EBF1, E2A and EBF1 act in concert to promote B cell development. EBF1 is a crucial factor in the regulatory circuitry that underlies B cell specification. EBF1 can restore the ability of PU.1-, Ikaros-, E2A-, and Pax5-deficient HPCs to differentiate into pro B cells that are committed to the B lineage and carry rearrangements of their immunoglobulin heavy-chain loci (IgH) (Seet et al. 2004; Kikuchi et al. 2005; Medina et al. 2004; Reynaud et al. 2008; Pongubala et al. 2008). Inactivation of EBF1 results in the complete block of B cell differentiation at the pre-pro B cell stage. Interestingly, EBF1-deficient HPCs also retain alternative lineage potential. The committed pro B cells with conditionally deleted Ebf1 acquire the potential to generate T cells and ILCs (Nechanitzky et al. 2013). Derepression of genes regulating T cell and ILC development, such as Id2 and TCF-1, was observed in the EBF1-deficient HPCs, suggesting an essential role of EBF1 in preventing the conversion to ILC or T cell fates by directly suppressing the expression of these target genes.
Pax5 has been also recognized as a master regulator of B cell commitment, as Pax5-deficient Pro B cells gain alternative lineage potential and dedifferentiate into non-B lineage cells (Nutt et al. 1999; Busslinger 2004). In addition, conditional deletion of Pax5 leads to reprogramming of mature B cells into T lineage cells (Cobaleda et al. 2007). Expression of non-B cell-associated genes, such as M-CFSR and Notch1, in Pax5-deficient Pro B cells suggests an essential role of Pax5 in suppressing genes involved in alternative lineage differentiation. Genome-wide analysis has identified the many Pax5 target genes in early and later stages of B cell development (Schebesta et al. 2007; Revilla et al. 2012).
5 Epigenetic Regulation in Lymphocyte Development
5.1 Polycomb Group Proteins and Lymphoid Lineage Commitment
Increasing evidence suggests that epigenetic factors play a crucial role in controlling the hematopoietic system. Among chromatin modifiers, Polycomb group (PcG) proteins are key regulators of lymphocyte cell fate (Zandi et al. 2010; Aloia et al. 2013). Several studies have elucidated the role of PcG proteins in HSC maintenance. One of the Polycomb repressive complexes (PRC) 1 components, Bmi1 has been reported to suppress the Ink4a/Arf locus, which encodes the cell cycle inhibitors p16 and p19 (Park et al. 2003). Moreover, Bmi1 is also implicated in repression of transcription factors essential for B cell determination, such as EBF1 and Pax5. Deletion of Bmi1 results in aberrant expression of EBF1 and Pax5, which leads to premature B lymphoid lineage specification (Oguro et al. 2010). In addition, Bmi1 was demonstrated to regulate thymocyte proliferation by suppressing the Ink4a/Arf locus (Miyazaki et al. 2008). Another PRC1 component, the Cbx family members, is also implicated in regulating self-renewal activities of HSCs, although the precise function seems to be different among the different Cbx genes (Klauke et al. 2013; van den Boom et al. 2013). Interestingly, Cbx4 has been reported to specifically regulate the proliferation and maintenance of thymic epithelial cells (Liu et al. 2013). Ring1B is a RING finger E3 ligase that monoubiquitylates H2A. Conditional deletion of Ring1B impairs B cell generation in BM, whereas the proliferation of immature progenitor cells and myeloid cells is enhanced (Cales et al. 2008). In addition of PRC1, PRC2 components have also been suggested to play an important role for proper HSC identity. Loss of Ezh2 severely impaired the self-renewal of fetal and adult HSCs (Mochizuki-Kashio et al. 2011; Hidalgo et al. 2012), and Ezh1-deficient mice have reduced number of HSCs in BM, suggesting some overlap, but not exactly same function of Ezh genes in the maintenance of HSC pools. Critical roles of Ezh2 in HSC self-renewal and early B cell development were also shown by inactivation of Ezh2 (Su et al. 2003; Kamminga et al. 2006). Taken together, these observations suggest that PcG proteins play a critical role at multiple stages in hematopoietic and lymphoid developmental system.
5.2 DNA Methylation and Other Chromatin Modifiers in Lymphoid Cell Fates
DNA methylation is an epigenetic mechanism essential for normal development, including transcriptional regulation, genomic imprinting, and genomic instability. DNA methyltransferases (Dnmts) are the group of enzymes responsible for establishment and maintenance of genomic DNA methylation. They include the de novo methyltransferases Dnmt3a and Dnmt3b and the maintenance methyltransferase Dnmt1. Conditional deletion of Dnmt1 in the hematopoietic system impairs the self-renewal of HSCs (Broske et al. 2009; Trowbridge et al. 2009). Dnmt3a−/−Dnmt3b−/− HSCs also have defects in their self-renewal activity, however, they can undergo normal differentiation to all hematopoietic lineages, indicating that de novo DNA methylation is not required for the differentiation of hematopoietic lineage cells (Tadokoro et al. 2007). Dnmt1 is also essential for proper development, survival, and function of T cells (Lee et al. 2001; Makar et al. 2003).
Special AT-Rich Sequence Binding protein 1 (SATB1) is a gene silencer that recruits chromatin-remodeling factors to regulate chromatin structure and gene expression. Expression of SATB1 increases during lymphoid lineage specification. Deletion of SATB1 reduces lymphopoietic activity of hematopoietic stem/progenitor cells, while the generation of myeloid lineage cells is unaffected. Ectopic expression of SATB1 promotes the differentiation toward T and B lymphoid lineages, suggesting a critical role of SATB1 in lymphoid lineage specification (Satoh et al. 2013).
Micro RNAs (miRNAs) are a recently discovered class of small (18-24 nt), noncoding RNAs that can downregulate target genes at the posttranscriptional level (Yuan and Muljo 2013). One miRNA, miR126, has recently been suggested to control B lineage specification independently of transcription factor activities. B cell but not myeloid development was promoted by the ectopic expression of miR126 in hematopoietic progenitors in fetal liver, both in vivo and in vitro. In addition, overexpression of miR126 in multipotent EBF1-deficient progenitors resulted in a partial induction of the B cell lineage program without upregulating E2A, EBF1, and Pax5 at the transcriptional level. These results indicate a critical role of miR126 in lymphoid versus myeloid lineage fates that is independent of canonical transcriptional cascades (Okuyama et al. 2013).
6 Transcriptional Regulation of ILC Development and the Relationship Between Acquired and Innate Lymphocytes
For many years, NK cells have been considered to be the only innate lymphocyte. However, it has recently become clear that innate lymphocytes are a more diverse group than previously anticipated. Innate lymphocytes, now called ILCs, are categorized into three groups, ILC1, ILC2, and ILC3 based on their functional characteristics, such as the cytokines that they produce and the transcription factors that are required for their development and function (Spits and Cupedo 2012; Walker et al. 2013). The generation of all ILCs depends on the transcriptional inhibitor Id2 (Klose et al. 2012). Id2 is one of a family of Id proteins that function as negative regulators of E proteins. Since E proteins, such as E2A and HEB normally promote T and B lymphocyte development, Id2 may function to induce ILC development by inhibiting adaptive lymphoid cell fate determination. ILCs also rely on the common cytokine receptor γ-chain (γc) and IL-7Rα, suggesting a critical role for IL-7 signaling in their development, similar to the situation with T and B lymphocyte development.
NK cells are a major component of ILC1 and are defined by the production of the IFNγ. CD122+NK1.1− NK cell-committed progenitors have been identified in fetal thymus and BM (Ikawa et al. 1999, 2001; Rosmaraki et al. 2001). Mice deficient for IL-2Rβ, IL-15Rα or IL-15 lack NK cells and detectable NK cell-mediated cytotoxicity, indicating a critical role of IL-15 signaling in the development and function of NK cells (Kennedy et al. 2000; Suzuki et al. 1997). The basic leucine zipper transcription factor E4BP4 was specifically shown to be required for the generation of NK cells. E4BP4-deficient mice lack NK cells. Real time RT-PCR analysis showed reduced expression of GATA3 and Id2 in E4BP4−/−HPCs in BM. Overexpression of Id2 in E4BP4−/−HPCs restored the production of NK cells, indicating that Id2 acts downstream of E4BP4 to promote NK cell development (Gascoyne et al. 2009).
ILCs that produce Th2-type cytokines IL-5 and IL-13 in response to stimulation by IL-25, IL-33, and thymic stromal lymphopoietin (TSLP) are classified as ILC2. ILC2 includes NH cells (Moro et al. 2010), nuocytes (Neill et al. 2010), type 2 innate helper (Ih2) cells, and multipotent progenitor type 2 (MPPtype2) cells (Price et al. 2010). Although their phenotypic markers are slightly different, the functional differences and relationships among these ILC2 populations remain to be determined. ILC2 requires the transcription factor retinoic acid receptor-related orphan receptor-α (RORα) (Halim et al. 2012; Wong et al. 2012) and GATA3 (Hoyler et al. 2012; Mjosberg et al. 2012; Furusawa et al. 2013) for their functions, such as antihelminth responses and allergic lung inflammation. About half of NH cells are marked with a history of Rag1 expression, detected using RAG1Cre/ROSA26YFP mice, implying a lymphoid developmental origin (Yang et al. 2011). This notion is also supported by the observation that the CLPs but not myeloid–erythroid progenitors gave rise to NH cells. In addition, Notch signaling has been shown to be essential for the generation of nuocytes from CLPs in vitro. As described above, TCF-1, which plays a critical role for T cell development, is also required for the development of ILC2 (Yang et al. 2013). Taken together, these observations indicate that T cells and ILC2 may share similar gene regulatory networks for their proper development and function.
ILC3 are defined by the capability to produce IL-17A and/or IL-22. The generation and function of ILC3 are totally dependent on the activity of transcription factor RORγt. On the basis of functional characteristics, ILC3 are categorized into three populations, LTi cells, IL-22-producing cells, and IL-17-producing cells. LTi cells were discovered almost two decades ago and represent the prototypic ILC3. LTi cells are crucial for the formation of secondary lymphoid organs during embryogenesis (van de Pavert and Mebius 2010). Although LTi cells appear to be closely related to other ILC3s, the relationship between them is still unclear. It also remains to be determined how the expression of RORγt is regulated during ILC3 development. More detailed characterization of each ILC3 subset and the relevant molecular mechanisms are needed for better understanding of the roles of these ILC populations.
7 Conclusions
Although many of the details regarding the mechanistic basis of lymphoid lineage determination by transcription factors and epigenetic factors have been elucidated, the exact structure of the network of these factors and the relationship among them remain largely unknown. Further dissection of the process between HSCs and committed T or B cells will be needed to draw an exact map of the network during lymphoid lineage specification. Establishment of in vitro models of lymphocyte development will also facilitate our understanding of the exact mechanisms of how these transcriptional networks operate. Combinations of genome-wide analysis, such as RNA-seq, ChIP-seq, and other epigenetic experimental systems will help us to fully clarify how cell-type specific gene programs are established and maintained to promote lymphocyte development. The development of the newly classified lymphoid cells, the ILCs, and the relationship between ILCs and T and B lymphocytes are intriguing issues to understand the overall development of the immune system.
References
Adolfsson J, Borge OJ, Bryder D, Theilgaard-Monch K, Astrand-Grundstrom I, Sitnicka E, Sasaki Y, Jacobsen SE (2001) Upregulation of Flt3 expression within the bone marrow Lin(−)Sca1(+)c-kit(+) stem cell compartment is accompanied by loss of self-renewal capacity. Immunity 15:659–669
Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, Bryder D, Yang L, Borge OJ, Thoren LA, Anderson K, Sitnicka E, Sasaki Y, Sigvardsson M, Jacobsen SE (2005) Identification of Flt3 + lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell 121:295–306
Agata Y, Tamaki N, Sakamoto S, Ikawa T, Masuda K, Kawamoto H, Murre C (2007) Regulation of T cell receptor beta gene rearrangements and allelic exclusion by the helix-loop-helix protein, E47. Immunity 27:871–884
Allman D, Sambandam A, Kim S, Miller JP, Pagan A, Well D, Meraz A, Bhandoola A (2003) Thymopoiesis independent of common lymphoid progenitors. Nat Immunol 4:168–174
Aloia L, di Stefano B, di Croce L (2013) Polycomb complexes in stem cells and embryonic development. Development 140:2525–2534
Balciunaite G, Ceredig R, Rolink AG (2005) The earliest subpopulation of mouse thymocytes contains potent T, significant macrophage, and natural killer cell but no B-lymphocyte potential. Blood 105:1930–1936
Bell JJ, Bhandoola A (2008) The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature 452:764–767
Broske AM, Vockentanz L, Kharazi S, Huska MR, Mancini E, Scheller M, Kuhl C, Enns A, Prinz M, Jaenisch R, Nerlov C, Leutz A, Andrade-Navarro MA, Jacobsen SE, Rosenbauer F (2009) DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet 41:1207–1215
Busslinger M (2004) Transcriptional control of early B cell development. Annu Rev Immunol 22:55–79
Cales C, Roman-Trufero M, Pavon L, Serrano I, Melgar T, Endoh M, Perez C, Koseki H, Vidal M (2008) Inactivation of the polycomb group protein Ring1B unveils an antiproliferative role in hematopoietic cell expansion and cooperation with tumorigenesis associated with Ink4a deletion. Mol Cell Biol 28:1018–1028
Christensen JL, Weissman IL (2001) Flk-2 is a marker in hematopoietic stem cell differentiation: a simple method to isolate long-term stem cells. Proc Natl Acad Sci USA 98:14541–14546
Cobaleda C, Jochum W, Busslinger M (2007) Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature 449:473–477
de Pooter RF, Kee BL (2010) E proteins and the regulation of early lymphocyte development. Immunol Rev 238:93–109
Dekoter RP, Singh H (2000) Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science 288:1439–1441
Dias S, Mansson R, Gurbuxani S, Sigvardsson M, Kee BL (2008) E2A proteins promote development of lymphoid-primed multipotent progenitors. Immunity 29:217–227
Durum SK (2003) Bcl11: sibling rivalry in lymphoid development. Nat Immunol 4:512–514
Furusawa J, Moro K, Motomura Y, Okamoto K, Zhu J, Takayanagi H, Kubo M, Koyasu S (2013) Critical role of p38 and GATA3 in natural helper cell function. J Immunol 191:1818–1826
Gascoyne DM, Long E, Veiga-Fernandes H, de Boer J, Williams O, Seddon B, Coles M, Kioussis D, Brady HJ (2009) The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat Immunol 10:1118–1124
Godfrey DI, Kennedy J, Suda T, Zlotnik A (1993) A developmental pathway involving four phenotypically and functionally distinct subsets of CD3-CD4-CD8—triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J Immunol 150:4244–4252
Halim TY, Maclaren A, Romanish MT, Gold MJ, McNagny KM, Takei F (2012) Retinoic-acid-receptor-related orphan nuclear receptor alpha is required for natural helper cell development and allergic inflammation. Immunity 37:463–474
Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K (1991) Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med 173:1213–1225
Hattori N, Kawamoto H, Fujimoto S, Kuno K, Katsura Y (1996) Involvement of transcription factors TCF-1 and GATA-3 in the initiation of the earliest step of T cell development in the thymus. J Exp Med 184:1137–1147
Hidalgo I, Herrera-Merchan A, Ligos JM, Carramolino L, Nunez J, Martinez F, Dominguez O, Torres M, Gonzalez S (2012) Ezh1 is required for hematopoietic stem cell maintenance and prevents senescence-like cell cycle arrest. Cell Stem Cell 11:649–662
Hosoya T, Kuroha T, Moriguchi T, Cummings D, Maillard I, Lim KC, Engel JD (2009) GATA-3 is required for early T lineage progenitor development. J Exp Med 206:2987–3000
Hoyler T, Klose CS, Souabni A, Turqueti-Neves A, Pfeifer D, Rawlins EL, Voehringer D, Busslinger M, Diefenbach A (2012) The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity 37:634–648
Igarashi H, Gregory SC, Yokota T, Sakaguchi N, Kincade PW (2002) Transcription from the RAG1 locus marks the earliest lymphocyte progenitors in bone marrow. Immunity 17:117–130
Ikawa T, Fujimoto S, Kawamoto H, Katsura Y, Yokota Y (2001) Commitment to natural killer cells requires the helix-loop-helix inhibitor Id2. Proc Natl Acad Sci USA 98:5164–5169
Ikawa T, Hirose S, Masuda K, Kakugawa K, Satoh R, Shibano-Satoh A, Kominami R, Katsura Y, Kawamoto H (2010) An essential developmental checkpoint for production of the T cell lineage. Science 329:93–96
Ikawa T, Kawamoto H, Fujimoto S, Katsura Y (1999) Commitment of common T/natural killer (NK) progenitors to unipotent T and NK progenitors in the murine fetal thymus revealed by a single progenitor assay. J Exp Med 190:1617–1626
Ikawa T, Kawamoto H, Goldrath AW, Murre C (2006) E proteins and notch signaling cooperate to promote T cell lineage specification and commitment. J Exp Med 203:1329–1342
Ikawa T, Kawamoto H, Wright LY, Murre C (2004) Long-term cultured E2A-deficient hematopoietic progenitor cells are pluripotent. Immunity 20:349–360
Ikuta K, Weissman IL (1992) Evidence that hematopoietic stem cells express mouse c-kit but do not depend on steel factor for their generation. Proc Natl Acad Sci USA 89:1502–1506
Inlay MA, Bhattacharya D, Sahoo D, Serwold T, Seita J, Karsunky H, Plevritis SK, Dill DL, Weissman IL (2009) Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes Dev 23:2376–2381
Kamminga LM, Bystrykh LV, de Boer A, Houwer S, Douma J, Weersing E, Dontje B, de Haan G (2006) The polycomb group gene Ezh2 prevents hematopoietic stem cell exhaustion. Blood 107:2170–2179
Kawamoto H, Ikawa T, Masuda K, Wada H, Katsura Y (2010) A map for lineage restriction of progenitors during hematopoiesis: the essence of the myeloid-based model. Immunol Rev 238:23–36
Kennedy MK, Glaccum M, Brown SN, Butz EA, Viney JL, Embers M, Matsuki N, Charrier K, Sedger L, Willis CR, Brasel K, Morrissey PJ, Stocking K, Schuh JC, Joyce S, Peschon JJ (2000) Reversible defects in natural killer and memory CD8 T cell lineages in interleukin 15-deficient mice. J Exp Med 191:771–780
Kikuchi K, Lai AY, Hsu CL, Kondo M (2005) IL-7 receptor signaling is necessary for stage transition in adult B cell development through up-regulation of EBF. J Exp Med 201:1197–1203
Klauke K, Radulovic V, Broekhuis M, Weersing E, Zwart E, Olthof S, Ritsema M, Bruggeman S, Wu X, Helin K, Bystrykh L, de Haan G (2013) Polycomb Cbx family members mediate the balance between haematopoietic stem cell self-renewal and differentiation. Nat Cell Biol 15:353–362
Klose CS, Hoyler T, Kiss EA, Tanriver Y, Diefenbach A (2012) Transcriptional control of innate lymphocyte fate decisions. Curr Opin Immunol 24:290–296
Kondo M, Weissman IL, Akashi K (1997) Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell 91:661–672
Kueh HY, Champhekhar A, Nutt SL, Elowitz MB, Rothenberg EV (2013) Positive feedback between PU.1 and the cell cycle controls myeloid differentiation. Science 341:670–673
Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, Cherry SR, Tsai JH, Tucker SM, Weaver WM, Kelso A, Jaenisch R, Wilson CB (2001) A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 15:763–774
Lee SU, Maeda M, Ishikawa Y, Li SM, Wilson A, Jubb AM, Sakurai N, Weng L, Fiorini E, Radtke F, Yan M, Macdonald HR, Chen CC, Maeda T (2013) LRF-mediated Dll4 repression in erythroblasts is necessary for hematopoietic stem cell maintenance. Blood 121:918–929
Li L, Leid M, Rothenberg EV (2010a) An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science 329:89–93
Li P, Burke S, Wang J, Chen X, Ortiz M, Lee SC, Lu D, Campos L, Goulding D, Ng BL, Dougan G, Huntly B, Gottgens B, Jenkins NA, Copeland NG, Colucci F, Liu P (2010b) Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science 329:85–89
Li YS, Wasserman R, Hayakawa K, Hardy RR (1996) Identification of the earliest B lineage stage in mouse bone marrow. Immunity 5:527–535
Liu B, Liu YF, Du YR, Mardaryev AN, Yang W, Chen H, Xu ZM, Xu CQ, Zhang XR, Botchkarev VA, Zhang Y, Xu GL (2013) Cbx4 regulates the proliferation of thymic epithelial cells and thymus function. Development 140:780–788
Liu P, Keller JR, Ortiz M, Tessarollo L, Rachel RA, Nakamura T, Jenkins NA, Copeland NG (2003) Bcl11a is essential for normal lymphoid development. Nat Immunol 4:525–532
Lu M, Tayu R, Ikawa T, Masuda K, Matsumoto I, Mugishima H, Kawamoto H, Katsura Y (2005) The earliest thymic progenitors in adults are restricted to T, NK, and dendritic cell lineage and have a potential to form more diverse TCRbeta chains than fetal progenitors. J Immunol 175:5848–5856
Lunardi A, Guarnerio J, Wang G, Maeda T, Pandolfi PP (2013) Role of LRF/pokemon in lineage fate decisions. Blood 121:2845–2853
Maeda T, Merghoub T, Hobbs RM, Dong L, Maeda M, Zakrzewski J, van den Brink MR, Zelent A, Shigematsu H, Akashi K, Teruya-Feldstein J, Cattoretti G, Pandolfi PP (2007) Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science 316:860–866
Makar KW, Perez-Melgosa M, Shnyreva M, Weaver WM, Fitzpatrick DR, Wilson CB (2003) Active recruitment of DNA methyltransferases regulates interleukin 4 in thymocytes and T cells. Nat Immunol 4:1183–1190
Mandel EM, Grosschedl R (2010) Transcription control of early B cell differentiation. Curr Opin Immunol 22:161–167
Mansson R, Zandi S, Welinder E, Tsapogas P, Sakaguchi N, Bryder D, Sigvardsson M (2010) Single-cell analysis of the common lymphoid progenitor compartment reveals functional and molecular heterogeneity. Blood 115:2601–2609
Massari ME, Murre C (2000) Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol 20:429–440
Masuda K, Kakugawa K, Nakayama T, Minato N, Katsura Y, Kawamoto H (2007) T cell lineage determination precedes the initiation of TCR beta gene rearrangement. J Immunol 179:3699–3706
Matsuzaki Y, Gyotoku J, Ogawa M, Nishikawa S, Katsura Y, Gachelin G, Nakauchi H (1993) Characterization of c-kit positive intrathymic stem cells that are restricted to lymphoid differentiation. J Exp Med 178:1283–1292
Medina KL, Pongubala JM, Reddy KL, Lancki DW, Dekoter R, Kieslinger M, Grosschedl R, Singh H (2004) Assembling a gene regulatory network for specification of the B cell fate. Dev Cell 7:607–617
Miyazaki M, Miyazaki K, Itoi M, Katoh Y, Guo Y, Kanno R, Katoh-Fukui Y, Honda H, Amagai T, van Lohuizen M, Kawamoto H, Kanno M (2008) Thymocyte proliferation induced by pre-T cell receptor signaling is maintained through polycomb gene product Bmi-1-mediated Cdkn2a repression. Immunity 28:231–245
Mjosberg J, Bernink J, Golebski K, Karrich JJ, Peters CP, Blom B, te Velde AA, Fokkens WJ, van Drunen CM, Spits H (2012) The transcription factor GATA3 is essential for the function of human type 2 innate lymphoid cells. Immunity 37:649–659
Mochizuki-Kashio M, Mishima Y, Miyagi S, Negishi M, Saraya A, Konuma T, Shinga J, Koseki H, Iwama A (2011) Dependency on the polycomb gene Ezh2 distinguishes fetal from adult hematopoietic stem cells. Blood 118:6553–6561
Moro K, Yamada T, Tanabe M, Takeuchi T, Ikawa T, Kawamoto H, Furusawa J, Ohtani M, Fujii H, Koyasu S (2010) Innate production of T(H)2 cytokines by adipose tissue-associated c-Kit(+)Sca-1(+) lymphoid cells. Nature 463:540–544
Nechanitzky R, Akbas D, Scherer S, Gyory I, Hoyler T, Ramamoorthy S, Diefenbach A, Grosschedl R (2013) Transcription factor EBF1 is essential for the maintenance of B cell identity and prevention of alternative fates in committed cells. Nat Immunol 14:867–875
Neill DR, Wong SH, Bellosi A, Flynn RJ, Daly M, Langford TK, Bucks C, Kane CM, Fallon PG, Pannell R, Jolin HE, McKenzie AN (2010) Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature 464:1367–1370
Nutt SL, Heavey B, Rolink AG, Busslinger M (1999) Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature 401:556–562
Nutt SL, Kee BL (2007) The transcriptional regulation of B cell lineage commitment. Immunity 26:715–725
Oguro H, Yuan J, Ichikawa H, Ikawa T, Yamazaki S, Kawamoto H, Nakauchi H, Iwama A (2010) Poised lineage specification in multipotential hematopoietic stem and progenitor cells by the polycomb protein Bmi1. Cell Stem Cell 6:279–286
Okuyama K, Ikawa T, Gentner B, Hozumi K, Harnprasopwat R, Lu J, Yamashita R, Ha D, Toyoshima T, Chanda B, Kawamata T, Yokoyama K, Wang S, Ando K, Lodish HF, Tojo A, Kawamoto H, Kotani A (2013) MicroRNA-126-mediated control of cell fate in B-cell myeloid progenitors as a potential alternative to transcriptional factors. Proc Natl Acad Sci USA 110:13410–13415
Osawa M, Hanada K, Hamada H, Nakauchi H (1996) Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science 273:242–245
Pai SY, Truitt ML, Ting CN, Leiden JM, Glimcher LH, Ho IC (2003) Critical roles for transcription factor GATA-3 in thymocyte development. Immunity 19:863–875
Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF (2003) Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423:302–305
Pongubala JM, Northrup DL, Lancki DW, Medina KL, Treiber T, Bertolino E, Thomas M, Grosschedl R, Allman D, Singh H (2008) Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat Immunol 9:203–215
Porritt HE, Rumfelt LL, Tabrizifard S, Schmitt TM, Zuniga-Pflucker JC, Petrie HT (2004) Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity 20:735–745
Price AE, Liang HE, Sullivan BM, Reinhardt RL, Eisley CJ, Erle DJ, Locksley RM (2010) Systemically dispersed innate IL-13-expressing cells in type 2 immunity. Proc Natl Acad Sci USA 107:11489–11494
Revilla IDR, Bilic I, Vilagos B, Tagoh H, Ebert A, Tamir IM, Smeenk L, Trupke J, Sommer A, Jaritz M, Busslinger M (2012) The B-cell identity factor Pax5 regulates distinct transcriptional programmes in early and late B lymphopoiesis. EMBO J 31:3130–3146
Reynaud D, Demarco IA, Reddy KL, Schjerven H, Bertolino E, Chen Z, Smale ST, Winandy S, Singh H (2008) Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat Immunol 9:927–936
Rosmaraki EE, Douagi I, Roth C, Colucci F, Cumano A, di Santo JP (2001) Identification of committed NK cell progenitors in adult murine bone marrow. Eur J Immunol 31:1900–1909
Rothenberg EV (2012) Transcriptional drivers of the T-cell lineage program. Curr Opin Immunol 24:132–138
Rothenberg EV, Moore JE, Yui MA (2008) Launching the T-cell-lineage developmental programme. Nat Rev Immunol 8:9–21
Satoh Y, Yokota T, Sudo T, Kondo M, Lai A, Kincade PW, Kouro T, Iida R, Kokame K, Miyata T, Habuchi Y, Matsui K, Tanaka H, Matsumura I, Oritani K, Kohwi-Shigematsu T, Kanakura Y (2013) The Satb1 protein directs hematopoietic stem cell differentiation toward lymphoid lineages. Immunity 38:1105–1115
Schebesta A, McManus S, Salvagiotto G, Delogu A, Busslinger GA, Busslinger M (2007) Transcription factor Pax5 activates the chromatin of key genes involved in B cell signaling, adhesion, migration, and immune function. Immunity 27:49–63
Schilham MW, Wilson A, Moerer P, Benaissa-Trouw BJ, Cumano A, Clevers HC (1998) Critical involvement of Tcf-1 in expansion of thymocytes. J Immunol 161:3984–3991
Schlenner SM, Madan V, Busch K, Tietz A, Laufle C, Costa C, Blum C, Fehling HJ, Rodewald HR (2010) Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity 32:426–436
Schwarz BA, Sambandam A, Maillard I, Harman BC, Love PE, Bhandoola A (2007) Selective thymus settling regulated by cytokine and chemokine receptors. J Immunol 178:2008–2017
Seet CS, Brumbaugh RL, Kee BL (2004) Early B cell factor promotes B lymphopoiesis with reduced interleukin 7 responsiveness in the absence of E2A. J Exp Med 199:1689–1700
Semerad CL, Mercer EM, Inlay MA, Weissman IL, Murre C (2009) E2A proteins maintain the hematopoietic stem cell pool and promote the maturation of myelolymphoid and myeloerythroid progenitors. Proc Natl Acad Sci USA 106:1930–1935
Spangrude GJ, Heimfeld S, Weissman IL (1988) Purification and characterization of mouse hematopoietic stem cells. Science 241:58–62
Spits H, Cupedo T (2012) Innate lymphoid cells: emerging insights in development, lineage relationships, and function. Annu Rev Immunol 30:647–675
Staal FJ, Luis TC, Tiemessen MM (2008) WNT signalling in the immune system: WNT is spreading its wings. Nat Rev Immunol 8:581–593
Staal FJ, Sen JM (2008) The canonical Wnt signaling pathway plays an important role in lymphopoiesis and hematopoiesis. Eur J Immunol 38:1788–1794
Su IH, Basavaraj A, Krutchinsky AN, Hobert O, Ullrich A, Chait BT, Tarakhovsky A (2003) Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat Immunol 4:124–131
Suzuki H, Duncan GS, Takimoto H, Mak TW (1997) Abnormal development of intestinal intraepithelial lymphocytes and peripheral natural killer cells in mice lacking the IL-2 receptor beta chain. J Exp Med 185:499–505
Tadokoro Y, Ema H, Okano M, Li E, Nakauchi H (2007) De novo DNA methyltransferase is essential for self-renewal, but not for differentiation, in hematopoietic stem cells. J Exp Med 204:715–722
Taghon T, Yui MA, Rothenberg EV (2007) Mast cell lineage diversion of T lineage precursors by the essential T cell transcription factor GATA-3. Nat Immunol 8:845–855
Ting CN, Olson MC, Barton KP, Leiden JM (1996) Transcription factor GATA-3 is required for development of the T-cell lineage. Nature 384:474–478
Trowbridge JJ, Snow JW, Kim J, Orkin SH (2009) DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell 5:442–449
van de Pavert SA, Mebius RE (2010) New insights into the development of lymphoid tissues. Nat Rev Immunol 10:664–674
van den Boom V, Rozenveld-Geugien M, Bonardi F, Malanga D, van Gosliga D, Heijink AM, Viglietto G, Morrone G, Fusetti F, Vellenga E, Schuringa JJ (2013) Nonredundant and locus-specific gene repression functions of PRC1 paralog family members in human hematopoietic stem/progenitor cells. Blood 121:2452–2461
Verbeek S, Izon D, Hofhuis F, Robanus-Maandag E, te Riele H, van de Wetering M, Oosterwegel M, Wilson A, Macdonald HR, Clevers H (1995) An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature 374:70–74
Wada H, Masuda K, Satoh R, Kakugawa K, Ikawa T, Katsura Y, Kawamoto H (2008) Adult T-cell progenitors retain myeloid potential. Nature 452:768–772
Walker JA, Barlow JL, McKenzie AN (2013) Innate lymphoid cells–how did we miss them? Nat Rev Immunol 13:75–87
Weber BN, Chi AW, Chavez A, Yashiro-Ohtani Y, Yang Q, Shestova O, Bhandoola A (2011) A critical role for TCF-1 in T-lineage specification and differentiation. Nature 476:63–68
Welinder E, Mansson R, Mercer EM, Bryder D, Sigvardsson M, Murre C (2011) The transcription factors E2A and HEB act in concert to induce the expression of FOXO1 in the common lymphoid progenitor. Proc Natl Acad Sci USA 108:17402–17407
Wong SH, Walker JA, Jolin HE, Drynan LF, Hams E, Camelo A, Barlow JL, Neill DR, Panova V, Koch U, Radtke F, Hardman CS, Hwang YY, Fallon PG, McKenzie AN (2012) Transcription factor RORalpha is critical for nuocyte development. Nat Immunol 13:229–236
Wu X, Satpathy AT, Kc W, Liu P, Murphy TL, Murphy KM (2013) Bcl11a controls Flt3 expression in early hematopoietic progenitors and is required for pDC development in vivo. PLoS One 8:e64800
Xu W, Carr T, Ramirez K, McGregor S, Sigvardsson M, Kee BL (2013) E2A transcription factors limit expression of Gata3 to facilitate T lymphocyte lineage commitment. Blood 121:1534–1542
Yamamoto R, Morita Y, Ooehara J, Hamanaka S, Onodera M, Rudolph KL, Ema H, Nakauchi H (2013) Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell 154:1112–1126
Yang L, Bryder D, Adolfsson J, Nygren J, Mansson R, Sigvardsson M, Jacobsen SE (2005) Identification of Lin(−)Sca1(+)kit(+)CD34(+)Flt3—short-term hematopoietic stem cells capable of rapidly reconstituting and rescuing myeloablated transplant recipients. Blood 105:2717–2723
Yang Q, Jeremiah Bell J, Bhandoola A (2010) T-cell lineage determination. Immunol Rev 238:12–22
Yang Q, Monticelli LA, Saenz SA, Chi AW, Sonnenberg GF, Tang J, de Obaldia ME, Bailis W, Bryson JL, Toscano K, Huang J, Haczku A, Pear WS, Artis D, Bhandoola A (2013) T cell factor 1 is required for group 2 innate lymphoid cell generation. Immunity 38:694–704
Yang Q, Saenz SA, Zlotoff DA, Artis D, Bhandoola A (2011) Cutting edge: natural helper cells derive from lymphoid progenitors. J Immunol 187:5505–5509
Yashiro-Ohtani Y, He Y, Ohtani T, Jones ME, Shestova O, Xu L, Fang TC, Chiang MY, Intlekofer AM, Blacklow SC, Zhuang Y, Pear WS (2009) Pre-TCR signaling inactivates notch1 transcription by antagonizing E2A. Genes Dev 23:1665–1676
Yu S, Zhou X, Steinke FC, Liu C, Chen SC, Zagorodna O, Jing X, Yokota Y, Meyerholz DK, Mullighan CG, Knudson CM, Zhao DM, Xue HH (2012) The TCF-1 and LEF-1 transcription factors have cooperative and opposing roles in T cell development and malignancy. Immunity 37:813–826
Yuan J, Muljo SA (2013) Exploring the RNA world in hematopoietic cells through the lens of RNA-binding proteins. Immunol Rev 253:290–303
Zandi S, Bryder D, Sigvardsson M (2010) Load and lock: the molecular mechanisms of B-lymphocyte commitment. Immunol Rev 238:47–62
Zheng W, Flavell RA (1997) The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 89:587–596
Zhu J, Min B, Hu-Li J, Watson CJ, Grinberg A, Wang Q, Killeen N, Urban JF Jr, Guo L, PAUL WE (2004) Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nat Immunol 5:1157–1165
Acknowledgments
We thank Peter Burrows for critical reading of the manuscript. This work is supported in part by grants from the Japan Society for the Promotion of Science (24689042), the Japan Science and Technology Agency, RIKEN IMS-RCAI Young Chief Investigator program and Kanae Foundation for the Promotion of Medical Science.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Ikawa, T. (2014). Genetic and Epigenetic Control of Early Lymphocyte Development. In: Ellmeier, W., Taniuchi, I. (eds) Transcriptional Control of Lineage Differentiation in Immune Cells. Current Topics in Microbiology and Immunology, vol 381. Springer, Cham. https://doi.org/10.1007/82_2014_370
Download citation
DOI: https://doi.org/10.1007/82_2014_370
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-07394-1
Online ISBN: 978-3-319-07395-8
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)