Abstract
Currently, regenerative medicine and cellular-based therapy have been in the center of attention worldwide in advanced medical technology. Mesenchymal stem cell (MSC) as a suitable stem cell source for cell-based therapy has been shown to be safe and effective in multiple clinical trial studies (CTSs) of several diseases. Despite the advantages, MSC needs more investigation to enhance its therapeutic application. The CRISPR/Cas system is a novel technique for editing of genes that is being explored as a means to improve MSCs therapeutic usage. In this study, we review the recent studies that explore CRISPR potency in gene engineering of MSCs, which have great relevance in MSC-based therapies. However, CRISPR/Cas technology make possible specific targeting of loci in target genes, but next-generation MSC-based therapies to achieve extensive clinical application need dedicated efforts.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keyword
1 Introduction
Cellular therapy has been defined as using healthy and effective cells for therapeutic purposes. However, is placed cell and tissue-based therapy together to introduce a new field of medicine that is called regenerative medicine. Cell and tissue-based therapies involve the transplantation of cells, tissues or their products developed for the purpose of repairing and/or restoring the function of diseased or dysfunctional cells or tissues. Therefore, there are different types of cells that are the candidate for using in cell therapy approach. These different types of cells can be categorized into three main groups include in stem cells, somatic cells, and genetically engineered cells (Golchin and Farahany 2019). As the introduction of regenerative medicine, the unique characteristics and potency of various source of stem cells have drawn a great deal of attention with many promises in the field of cell-tissue based therapy (Golchin et al. 2019). Among different source of stem cells, mesenchymal stem cells (MSCs) due to their suitable features and accessibility have been more commonly used in cell-based therapy research and clinical applications (Golchin et al. 2018a).
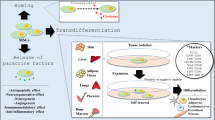
Gene therapy, that provides an innovative treatment option, is defined as introducing genetic material into living cells to compensate for abnormal genes or to express a beneficial protein for treating or preventing of certain diseases (Kohlscheen et al. 2017; Golchin and Farahany 2019). Transfer of gene-corrected auto/allogeneic stem cells in some patients has emerged as a new therapeutic approach. As mentioned, MSCs are primordial, unspecialized and undifferentiated cells containing the potential of self-renewal through continuous cell division and differentiation into various other types of cells We discussed the MSCs underlying advantages and limitations and reviewed the genetically engineering guideline for clinical MSC therapy to improve their therapeutic efficacy in a separate study (Golchin et al. 2018b). In order to overcome the technical challenge of MSCs for therapeutic applications, gene engineering provides several gene editing systems include meganucleases, zinc-finger nucleases (ZNFs) system, Transcription activator-like effector nucleases (TALEN) system and clustered regularly interspaced short palindromic repeats (CRISPR)-associated-9 (CRISPR/Cas9 system). In this study, we focused on CRISPR/Cas9-engineered MSCs (Fig. 1) as a new and effective tool for developing cell-based therapy.
2 Clinical Advantages of MSCs in Regenerative Medicine
Mesenchymal stem cells (MSCs) are used in many types of research because of self-renewing and multipotent adult stem cells of mesodermal origin with a considerable potency to differentiate into several cell types like chondrocytes, adipocytes, osteoblasts, and other cell types (Ardeshirylajimi et al. 2014, 2017). As mesenchymal stem cells (MSCs) reside are placed mainly within the stromal portion of bone marrow, have multiple differentiation potentials under appropriate conditions (Shen et al. 2018). The International Society for Cellular Therapy (ISCT), as a global society with a shared vision to translate cellular therapy into safe and effective therapies, listed the minimum criteria and markers of MSCs that include (Dominici et al. 2006):
-
1.
Plastic-adherent cells isolated from different tissues in the standard culture conditions;
-
2.
Specific surface antigen (Ag) expression: positive expression for CD105, CD73, CD90, and negative for markers including CD45, CD34, CD14 or CD11b, CD79α, CD19, and HLA-DR;
-
3.
In vitro differentiation into three cell types including osteoblasts, adipocytes, and chondrocytes.
There are several special advantages for MSCs in comparison other stem cells, for instance, lack of their ethical issue, easily accessible and isolated from different tissues (such as bone marrow (Friedenstein et al. 1987), adipose tissues (Zuk et al. 2002), umbilical cord (McElreavey et al. 1991) and etc), suitable differentiation potential (differentiated to adipocytes, osteoblasts, chondrocytes, myoblasts, and etc. (Chamberlain et al. 2007)), good proliferation rate, and safety for clinical application (Golchin et al. 2018a). MSCs can migrate to the injury sites and carry out immune regulation, site-specific differentiation, support hematopoiesis (Ullah et al. 2015). Therefore, they are perfect candida ting cells in widely applied in experimental and clinical researches and gene engineering for regeneration of bone, heart, cartilage, central nervous, skin and so on that possessing a great application landscape in the field of tissue repair (Reiser et al. 2005).
As mentioned, MSCs are present in several tissues such as liver, skin, bone marrow, dental pulp, brain, adipose tissue and skeletal muscle and are associated in processes like immunosuppression and have an ability to migrate towards sites of tumors and inflammation zones. Hence, owing to their differentiation capabilities, easily isolation, and immunomodulatory features, the therapeutic potential of mesenchymal stem cells (MSCs) has been determined in many pre-clinical and clinical settings (Zhang et al. 2017). All of these reasons and most importantly high self-renewal potential makes them a great candidate for delivering genes and restituting organ systems function (Shen et al. 2018).
Alongside advantages of MSCs, there are several limitations that decrease the efficacy of therapeutic properties of MSCs. For instance, the low potency in biological (in vivo) condition comparison in vitro condition (Samsonraj et al. 2015), the low homing rate in the target site, insufficient expression of some factors and low cell viability after transplantation (Golchin et al. 2018b). Forasmuch as gene therapy and gene-engineering allow the addition of new functions to cells, this opportunity is provided to enhance MSCs features and applications.
3 Gene-Engineering for Stem Cell Therapy
In recent years, the appearance of varied genome-editing technologies has provided the ability to economically and rapidly introduce sequence-specific modifications into the genomes of a wide range of cell types for biologist and researchers (Gaj et al. 2016). For this purpose, different methods such as physical and chemical non-viral methods and viral vector-based methods are used to introduce target genes to MSCs.
-
Non-viral method: Physical (Electroporation, microinjection, plasmid-injection, Ballistic injection) and Chemical (Liposome-based methods, calcium phosphate, DEAE dextran, protein-based methods)
-
Viral methods: RNA virus (Retrovirus, HIV (lentivirus) and DNA virus (Adenovirus, Adeno-associated virus (AAV), Herpes simplex virus)
Genome editing with programmable nucleases has opened a new way for various applications from basic research in disease model via animal and cellular models to regenerative medicine and clinical trial studies (Barrangou et al. 2007). A series of studies showed that genome editing could greatly stimulate by targeted DNA double-strand breaks (DSBs). Till now for genome editing, four major classes of adjustable DNA-binding proteins have been engineered based on site-specific DNA DSBs: meganucleases or homing endonucleases obtained from microbial mobile genetic components, zinc finger (ZF) nucleases based on eukaryotic transcription factors, transcription activator-like effectors (TALEs) from Xanthomonas bacteria, and the RNA-guided DNA endonuclease Cas9 from the type II bacterial adaptive immune system CRISPR that found recently (Cong et al. 2013). Genome editing based on nuclease systems can be classified into two groups via their mode of DNA identification- TALEN, ZFN, and meganucleases attain specific DNA binding by protein-DNA interactions. While Cas9 is targeted to particular DNA sequences by a short RNA guide molecule and after that its targeting DNA, protein-DNA interactions that have an important role (Bayes-Genis et al. 2005). The modification of MSCs properties is necessary to fully use their potential. Gene-engineering with novel techniques to induce gene expression in a correct and considerable manner is particularly attractive for stem cell-based therapy purpose.
4 Meganucleases
The LAGLIDADG family is the largest class of homing endonucleases, which contains the well-characterized and generally used I-CreI and I-SceI enzymes. They are the smallest class of engineered nucleases with large (>14 bp) recognition sites that help them potentially amenable to all standard gene delivery methods (Bitinaite et al. 1998). Although many studies suggest using meganucleases in genome editing, an important problem was reported about cleavage domains of endonucleases and the DNA-binding that are difficult to separate. For solving this limitation, chimeric proteins consist of ZFs, meganucleases and TALEs have been engineered to generate novel monomeric enzymes. Formation of DSB by these enzymes results in a 3′ overhang that can be more recombinogenic for HDR than 5′ overhang generated by FokI cleavage and this is one of the meganuclease technology advantages. So, multiple meganuclease monomers could be wrapped into single viral vectors to make multiple DSBs simultaneously (Bitinaite et al. 1998).
5 Zinc Finger
Zinc finger (ZF) proteins are the large class of transcription factors and the Cys2-His2 zinc finger domain is one of the most current DNA-binding domains encoded in the human genome. By detection the independent function of the DNA-binding domain and the cleavage domain of the FokI restriction endonuclease, the zinc finger nuclease (ZFN) technology was made. As the FokI nuclease acting as a dimer, using two ZFNs binding opposite strands of DNA are needed for induction of a DSB. Since ZFN-induced DSBs were used to modify the genome through either NHEJ or HDR, this technology has been applied to modify genes in a pluripotent stem and human somatic cells successfully (Sebastiano et al. 2011). One of the great concern connected with the use of ZFNs for genome editing is off-target mutations (Koo et al. 2015).
6 TALEN
The development of TALEN system is associated with the study of the Xanthomonas genus bacteria that secrete effector proteins (transcription activator-like effectors) via capable of DNA binding and activating the expression of their target genes by mimicking the eukaryotic transcription factors (Hockemeyer et al. 2011). Like ZFNs, TALENs consist of individual modules targeting 3 or 1 nucleotides (nt) of DNA, respectively. Also like ZFNs, TALENs are modular in form and dimerization of TALEN proteins is mediated by the FokI cleavage domain, which cuts within a 12- to 19-bp spacer sequence that detaches each TALE binding site. The DNA-binding domain was indicated to contain monomers; each of them binds one nucleotide in the target nucleotide sequence. Monomers are tandem repeats of 34 amino acid residues, two of which are highly variable located at 12 and 13 positions, and they are responsible for the diagnosis of a particular nucleotide (Nemudryi et al. 2014). Thymidine is the target DNA molecule that affects the binding efficiency and locates before the5’-end of a sequence bound by a TALE monomer. A half-repeat is the last tandem repeat that binds a nucleotide at the 3′-end of the diagnosis site consists only of 20 amino acid residues. There are two distinct advantages for TALENs compared with ZFNs in genome editing I: they have been reported to indicate ameliorated specificity and decreased toxicity compared to some ZFNs, because of their increased affinity for target DNA.II: There is no selection or directed evolution for engineering TALE arrays, so reducing the amount of time and experience that needed to collect a functional nuclease (Maeder and Gersbach 2016). The absence of obvious correspondence between meganuclease protein residues and their target DNA sequence caused that meganucleases have not been adopted as a genome engineering platform. On the other hand, ZF domains, because of interference between neighbor modules when gathered into a larger array, exhibit context-dependent binding priority. Identically, although TALE DNA-binding monomers are for the most part modular, they can travail from context-dependent specificity and their repetitive sequences provide a construction of novel TALE arrays labor intensive and expensive (Hsu et al. 2014).
Multiple strategies have been developed to account for these limitations one of them was CRISPR nuclease Cas9. Because the Cas9 protein is constant and can be retargeted to new DNA sequences easily by changing a small portion of the sequence of an accompanying RNA guide that base-pairs with target DNA straightly. Also, an important potential of Cas9 is its ability to demonstrate multiple DSBs in the same cell via expression of separate guide RNAs (Cong et al. 2013).
7 CRISPR/Cas Nucleases
CRISPR (clustered regularly interspaced short palindromic repeat)-Cas RNA-guided nucleases are derived from an adaptive immune system that progressed in bacteria for preventing assault of viruses, plasmids and exogenous genetic elements (EGEs) that incorporate with Cas proteins. After a decades, scientists could illuminate a mechanism that short sequences of invading nucleic acids were consolidated into CRISPR loci (Maeder and Gersbach 2016). Then these sequences transcribe and process three main components to cleave foreign nucleic acids CRISPR RNAs (crRNAs), trans-activating crRNAs (tracrRNAs) and CRISPR associated (Cas9) endonuclease. crRNA sequences are complementary to exogenous genetic elements and acting as a target site-specific sequences which will be cleaved by the Cas9 endonucleases. The tracrRNA has homology regions and acts as a link between the variable crRNAs and Cas9. For simplifying laboratory applications the crRNA and tracrRNA have been composed into a single chimeric RNA sequence named short guide RNA (sgRNA) (Albitar et al. 2018). In order to ensure DNA diagnosis and cleavage, six CRISPR systems according to different mechanisms have been identified. These systems are divided into two classes: Class 1(types I, III, and IV), and Class 2 (types II, V, and VI). Class 2 systems due to their simplicity were appealing for genome engineering and only type II that obtained from Streptococcus pyogenes,has been used for RNA-guided engineered nucleases (Koonin et al. 2017). The effector protein of type II CRISPR-Cas systems is Cas9 and this multi-task protein has been engineered into a key tool for genome editing. The guide RNA (sgRNA) manages the CRISPR associated protein Cas9 duo to present a sequence specific DNA cleavage by double-strand breaks (DSBs) in the target DNA. NGG motif or proto-spacer adjacent motif (PAM) is a short-conserved sequence that is required for introducing a break. Hence, CRISPR/Cas9 by utilizing sgRNA with Cas9 nuclease can recognize a variable 20-nucleotide target sequence adjacent to a 5’-NGG-3’ PAM and introduces a DSB in the target DNA three base pairs upstream of the PAM sequence (Li et al. 2018a, b). Since the induced DSB is a lethal happening for cells, these cells need a mechanism for DNA repair. These mechanisms consist of the homology-direct repair (HDR) pathways and the non-homologous end joining (NHEJ). The HDR pathway of DNA damage repair includes a precise strand-exchange process based on existing homologous DNA formats, which contain homology to sequences flanking the DSB demonstrated by homology arms (He et al. 2016). The mechanism of NHEJ repair consists in joining of the free DNA ends via a homology independent and mechanistically flexible process, which often produces random small deletions or insertions (Albitar et al. 2018).
8 Advantages of CRISPR
The CRISPR/Cas9 genome-editing system proposes several advantages over transcription activator like effector nuclease (TALEN) and the zinc-finger nucleases (ZFNs) in adult stem cells (ASCs) and human pluripotent stem cells (PSCs). First of all, CRISPR/Cas9 is more economical because there is little related cost for plasmid-mediated CRISPR/Cas9. Second, as the fastest existing genome-editing technique, because this system can typically be performed in 2 weeks. Third, CRISPR/Cas9 is more user-friendly than TALEN and ZNF (Zhang et al. 2017). Fourth, the capability of Cas9 to display multiple DSBs in the same cell via expression of separate guide RNAs is a potential advantage. At last, CRISPR/Cas9 displays a higher editing efficiency than TALEN and ZNF in human stem cells. CRISPR can target multiple loci simultaneously in the genome with high efficiency and without remarkably increasing the required dose. As XL et al. demonstrated treatment with a BCL inhibitor ABT-263 further improves HDR efficiency by 70% and knockout (KO) efficiency by 40% via CRISPR-Cas9 in human pluripotent stem cells. The increased efficiency of genome editing is ascribed to higher expressions of Cas9 and sgRNA in surviving cells after electroporation (Li et al. 2018a, b). Table 1 demonstrates comparison of different programmable nuclease platforms. However, CRISPR/Cas9 technology is one of the great promises as a means to produce biological products and especially therapeutic cellular products.
9 Application of CRISPR/Cas9 in Mesenchymal Stem Cell Studies
CRISPR/Cas9-based gene manipulation including in gene knock-in, gene knockout, gene activation or interference, and other chromosome-related usages, has been widely employed in stem cell research and specially MSC research (Table 2; Shen et al. 2018).
One of the main limitations of cell therapy is the immune rejection of transplanted cells. Due to no or low expression of MHC class II proteins, MSCs prevent allogeneic rejection. However, studies don’t refuse the role of MHC class 1 in immune rejection completely (Fukami et al. 2009; Ayala García et al. 2012). The result of a study has reported that hMSC with B2M (the light chain of MHC class I molecule (Chen et al. 2017)) knockdown by CRISPR/Cas9 is a suitable and useful stem cell source to treat myocardial infarction without inducing immune rejection (Li et al. 2018b). Another study has reported that by knocking-out β2-microglobulin (B2M) in primary hMSCs can be utilized to increase the gene ablation rate in cells relevant to clinical applications (Xu et al. 2018).
Both of viral and non-viral vectors could be used in CRISPR/Cas9-engineered MSCs (Meca-Cortés et al. 2017; Xu et al. 2018). Meca-Cortés et al. report that the therapeutic capacity of the electroporation as a transfection method for CRISPR/Cas9-engineered hAMSCs is equivalent to that of therapeutic hAMSCs generated by introduction of the same therapeutic gene by transduction with a lentivirus vector (Meca-Cortés et al. 2017).
In recent years, the use of MSCs in both gene and cell therapies especially as vehicles has accelerated. For example, MSCs can be used as vehicles to deliver anti-tumor agents and drugs to tumor sites. Almeida demonstrated the genetic edition of MSCs to be vehicles for drug delivery of azurin into target sites due to their migration potential towards tumors and unique immunomodulation. The primary steps of this strategy were the designing and testing of gRNAs to produce DSBs in a genomic safe harbor, and the design of a donor pattern that causes the interpolation of the azurin gene that encodes this protein into safe locus via CRISPR/Cas9 (Filipa and Almeida 2017). According to azurin properties as an anticancer protein and the tropism ability of the MSCs towards tumor sites, the formulated strategy of this work was to test the possibility of steadily incorporating a gene coding for azurin within the genome of MSCs. In this study, the Cas9 guides were tested in MSCs and HEK293T cells and selecting one guide for CRISPR/Cas9 technology in order to cleavage the selected safe harbor AAVS1 locus in the intron 1 of PPP1R12C gene was done. After the design of the guides, these were tested in HEK293T cells displaying that the Guide 3 was also the best considering its cleavage efficiency observed in the agarose gel. The best guide was Guide 3 because of its good score and the zero exonic off-targets was tested in the second cell line (MSCs). The producers of azurin in MSCs was not done in this step because of problems in designing the guides RNA and surveying the off-targets while these items were done successfully. Hence, the next steps are the ligations between azurin gene AAVS1 locus to produce a donor template capable of repairing the DSB by using MSCs in future experiments (Filipa and Almeida 2017).
Considering the importance of mesenchymal stem cells (MSCs) for curing type 1 diabetes (T1D), Gerace et al. suggested utilization of clustered regularly interspaced short palindromic repeat (CRISPR) for performing the improved clinical trial design for the future success of T1D MSC derived therapies. Although islet or pancreas transplantation is the only cure for people with type 1 diabetes (T1D), MSCs have been employed either natively or transdifferentiated into insulin-producing cells (IPCs) as a second treatment (Gerace et al. 2017). As some researches showing the ability of MSCs to differentiate into insulin-producing cells (IPCs) via ex vivo chemical induction or different gene therapy procedures describes them as ideal candidates for cell transplantation. Gerace and colleague displayed the success of MSC-derived therapies in pre-clinical models and reflected the failure of the translation of these studies into the clinical setting. Hence, the limitations of common clinical trials of MSCs for the treatment of T1D suggested the novel clustered regularly interspaced short palindromic repeat (CRISPR) gene-editing technology for ameliorating the clinical trial plan as strategies to translate pre-clinical success to the clinical setting (Gerace et al. 2017).
Another study done by Shen et al. was about gene editing of PTEN in MSCs and its changes in differentiation and proliferation in vivo (Shen et al. 2018). As the tumor suppressor, PTEN is associated with lineage determination, motility, the regulation of cell proliferation, apoptosis and adhesion. Mutation or loss of PTEN has existed in several human cancers and diverse hereditary disorders. Since PTEN was recognized to increase MSCs migration ability, this study clarifies the role of PTEN in the in vivo proliferation and differentiation via a gene-editing approach. They used CRISPR/Cas9 to knockout the PTEN gene in MSCs and obtained the PTEN-KO BMSCs from rabbit. Results illustrated that rabbit BMSCs are agreeable to accurate genetic manipulations. By using this technology for PTEN knockout cells, increased proliferation capability and decreased osteogenic and adipogenic differentiation ability was shown compared with the WT. These results display when BMSCs using as the seed cells for tissue engineering, indicated a low expression of PTEN, the findings suggest a spoiled differentiation and tissue repair function (Shen et al. 2018). Recently, a study was done by van den Akker and colleague about CRISPR/Cas9 technique for inactivation of genes in hMSC (van den Akker et al. 2016). They determined the possibility of generating knock-out cell populations from human mesenchymal stem cells, without sub-cloning of cells. As transforming Growth Factor (TGF-ß) signaling is important for chondrogenic differentiation of MSC and the conservation of the articular chondrocyte phenotype, CRISPR guide RNAs were designed to target the second coding exon of the five R-SMAD genes and cloned into a lentiviral Cas9 expression system. The efficiency of CRISPR was evaluated by using surveyor nuclease assay on MSC and HEK293 cells. The surveyor nuclease assay displayed a higher percentage of genomic modification. This targeting strategy reduced SMAD protein expression in HEK293T by 90% and MSC was expected to be more unprotected to CRISPR/Cas9 genome engineering or high viral loads. Hence, primary findings determine that MSC expressing CRISPR/Cas9 steadily show normal differentiation characteristics (van den Akker et al. 2016). All these researches are in the early stages and required more time for finding more acceptable results and using them in clinical trials.
10 Conclusion and Outlook
The rise of powerful, effective and cost-effective methods for the genetic manipulation of cells is opening up wide prospects for cell-based therapy. CRISPR/Cas has emerged as future technologies due to the rapidity and specificity of gene delivery using gene-editing techniques. MSCs as an accessible and suitable source of stem cell confirmed by extensive research. According to preclinical and clinical trial studies, MSCs considered nearby to approved clinical therapeutics (Golchin et al. 2018a). On the other hand, the ability to modify a cell’s DNA with precision and achieve of gene-engineered cells in biomedicine, enabled by methods based on CRISPR, has paved the way for a degree of appropriate cell customization for clinical application. Therefore, combining the stemness potential of MSCs with CRISPR/Cas9 technology has made to an interesting field that made an accessible tool to clinical application. Despite different research for using mesenchymal stem cells (MSCs) via (CRISPR) gene-editing has been registered only one in clinical trials in this regard. Currently, a clinical trial is registered in clinicaltrial.gov that combine the self-renewal potential of MSCs with CRISPR/Cas9 technology that will be developed an epigenome editing approach as a therapeutic strategy to rescue the activity of MLL4 (51). It is expected that the number of clinical trials in this regards enhances. However, CRISPR/Cas technology enables specific targeting of loci in target genes, but next-generation stem cell-based therapies to achieve widespread application need dedicated efforts.
Abbreviations
- CRISPR/Cas9:
-
Clustered Regularly Interspaced Short Palindromic Repeats-associated-9
- crRNAs:
-
CRISPR RNAs
- CTSs:
-
Clinical Trial Studies
- DSBs:
-
Double-Strand Breaks
- hESC:
-
human Embryonic Stem Cell
- iPS:
-
Induced Pluripotent Stem Cells
- IVF:
-
in vitro Fertilization
- MSC:
-
Mesenchymal Stem Cell
- PAM:
-
Proto-spacer Adjacent Motif
- SCNT:
-
Somatic-cell Nuclear Transfer
- TALEN:
-
Transcription Activator-like Effector Nucleases
- tracrRNAs:
-
Trans-activating crRNAs
- ZNFs:
-
Zinc-Finger nucleases
References
Albitar A, Rohani B, Will B et al (2018) The application of CRISPR/Cas technology to efficiently model complex Cancer genomes in stem cells. J Cell Biochem 119:134–140. https://doi.org/10.1002/jcb.26195
Ardeshirylajimi A, Soleimani M, Hosseinkhani S et al (2014) A comparative study of osteogenic differentiation human induced pluripotent stem cells and adipose tissue derived mesenchymal stem cells. Cell J 16:235
Ardeshirylajimi A, Vakilian S, Salehi M, Mossahebi-Mohammadi M (2017) Renal differentiation of mesenchymal stem cells seeded on nanofibrous scaffolds improved by human renal tubular cell lines-conditioned medium. ASAIO J 63:356–363. https://doi.org/10.1097/MAT.0000000000000470
Ayala García MA, González Yebra B, López Flores AL, Guaní Guerra E (2012) The major histocompatibility complex in transplantation. J Transp Secur 2012:842141–842147. https://doi.org/10.1155/2012/842141
Barrangou R, Fremaux C, Deveau H et al (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science (80- ) 315:1709–1712. https://doi.org/10.1126/science.1138140
Bayes-Genis A, Roura S, Soler-Botija C et al (2005) Identification of cardiomyogenic lineage markers in untreated human bone marrow–derived Mesenchymal stem cells. Transplant Proc 37:4077–4079. https://doi.org/10.1016/j.transproceed.2005.09.103
Bitinaite J, Wah DA, Aggarwal AK et al (1998) FokI dimerization is required for DNA cleavage. Proc Natl Acad Sci 95:10570–10575. https://doi.org/10.1073/pnas.95.18.10570
Carpenter MK, Rao MS, Carpenter MKRM et al (2015) Concise review: making and using clinically compliant pluripotent stem cell lines. Stem Cells Transl Med 4:381–388. https://doi.org/10.5966/sctm.2014-0202
Carstairs A (2017) Development of in vitro skeletal disease models using CRISPR/Cas9 genome editing in immortalised mesenchymal stem cells
Chamberlain G, Fox J, Ashton B, Middleton J (2007) Concise review: mesenchymal stem cells: their phenotype, differentiation capacity, immunological features, and potential for homing. Stem Cells 25:2739–2749. https://doi.org/10.1634/stemcells.2007-0197
Chen Z, Zhang N, Qi J et al (2017) The structure of the MHC class I molecule of bony fishes provides insights into the conserved nature of the antigen-presenting system. J Immunol 199:3668–3678. https://doi.org/10.4049/jimmunol.1600229
Cong L, Ran FA, Cox D et al (2013) Multiplex genome engineering using CRISPR/Cas systems. Science (80-) 339:819–823. https://doi.org/10.1126/science.1231143
Dominici M, Le Blanc K, Mueller I et al (2006) Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy Position Statement Cytotherapy 8:315–317. https://doi.org/10.1080/14653240600855905
Filipa A, Almeida D (2017) Genome editing of Mesenchymal stem / stromal cells (MSCs) by CRISPR/Cas9 technology for azurin-based anticancer therapies. Tec LISBOA:1–11
Friedenstein AJ, Chailakhyan RK, Gerasimov UV (1987) Bone marrow osteogenic stem cells: in vitro cultivation and transplantation in diffusion chambers. Cell Tissue Kinet 20:263–272
Fukami N, Ramachandran S, Saini D et al (2009) Antibodies to MHC class I induce autoimmunity: role in the pathogenesis of chronic rejection. J Immunol 182:309–318
Furuhata Y, Nihongaki Y, Sato M, Yoshimoto K (2017) Control of adipogenic differentiation in mesenchymal stem cells via endogenous gene activation using CRISPR-Cas9. ACS Synth Biol 6:2191–2197. https://doi.org/10.1021/acssynbio.7b00246
Gaj T, Sirk SJ, Shui S, Liu J (2016) Genome-editing technologies: principles and applications. Cold Spring Harb Perspect Biol 8:a023754. https://doi.org/10.1101/cshperspect.a023754
Gerace D, Martiniello-Wilks R, Nassif NT et al (2017) CRISPR-targeted genome editing of mesenchymal stem cell-derived therapies for type 1 diabetes: a path to clinical success? Stem Cell Res Ther 8:62. https://doi.org/10.1186/s13287-017-0511-8
Golchin A, Farahany TZ (2019) Biological products: cellular therapy and FDA approved products. Stem Cell Rev Rep 15:1–10. https://doi.org/10.1007/s12015-018-9866-1
Golchin A, Farahany TZ, Khojasteh A et al (2018a) The clinical trials of mesenchymal stem cell therapy in skin diseases: an update and concise review. Curr Stem Cell Res Ther 13:22–33. https://doi.org/10.2174/1574888X13666180913123424
Golchin A, Rekabgardan M, Taheri RA, Nourani MR (2018b) Promotion of cell-based therapy: special focus on the cooperation of mesenchymal stem cell therapy and gene therapy for clinical trial studies. In: Turksen K (ed) Advances in experimental medicine and biology. Springer, New York, pp 103–118
Golchin A, Shams F, Kangari P et al (2019) Regenerative medicine: injectable cell-based therapeutics and approved products. Adv Exp Med Biol. Springer, New York, pp 1–21
He X, Tan C, Wang F et al (2016) Knock-in of large reporter genes in human cells via CRISPR/Cas9-induced homology-dependent and independent DNA repair. Nucleic Acids Res 44:e85–e85. https://doi.org/10.1093/nar/gkw064
Hockemeyer D, Wang H, Kiani S et al (2011) Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol 29:731–734. https://doi.org/10.1038/nbt.1927
Hsu PD, Lander ES, Zhang F (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell 157:1262–1278. https://doi.org/10.1016/j.cell.2014.05.010
Hu X, Li L, Yu X et al (2017) CRISPR/Cas9-mediated reversibly immortalized mouse bone marrow stromal stem cells (BMSCs) retain multipotent features of mesenchymal stem cells (MSCs). Oncotarget 8:111847–111865. https://doi.org/10.18632/oncotarget.22915
Kohlscheen S, Bonig H, Modlich U (2017) Promises and challenges in hematopoietic stem cell gene therapy. Hum Gene Ther 28:782–799. https://doi.org/10.1089/hum.2017.141
Koo T, Lee J, Kim J-S (2015) Measuring and reducing off-target activities of programmable nucleases including CRISPR-Cas9. Mol Cells 38:475–481. https://doi.org/10.14348/molcells.2015.0103
Koonin EV, Makarova KS, Zhang F (2017) Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol 37:67–78. https://doi.org/10.1016/j.mib.2017.05.008
Kosaric N, Srifa W, Gurtner GC, Porteus MH (2017) Human Mesenchymal stromal cells engineered to overexpress PDGF-B using CRISPR/Cas9/rAAV6-based tools improve wound healing. Plast Reconstr Surg – Glob Open 5:74. https://doi.org/10.1097/01.GOX.0000516619.11665.84
Li C, Yue S, Busuttil R, Kupiec J, Weglinski BK (2016) Mesenchymal stem cells reprogram macrophage polarization and attenuate liver ischemia and reperfusion injury by regulating notch/Cox2/PGE2 signaling pathway. ATC Abstracts. https://atcmeetingabstracts.com/abstract/mesenchymal-stem-cells-reprogram-macrophage-polarization-and-attenuate-liver-ischemia-and-reperfusion-injury-by-regulating-notchcox2pge2-signaling-pathway/. Accessed 20 Mar 2019
Li X-L, Li G-H, Fu J et al (2018a) Highly efficient genome editing via CRISPR–Cas9 in human pluripotent stem cells is achieved by transient BCL-XL overexpression. Nucleic Acids Res 46:10195–10215. https://doi.org/10.1093/nar/gky804
Li Y, Shao L, Yu Z, Wang J, Wang Y, Yuqing Zhang CH, Zhu B, Zhang L, Pan X, Ouyang W, Liu B, Liang C, Geng Y-j, X yong Y (2018b) Downregulation of B2M in allogeneic MSC by CRISPR technology inhibits immune rejection and enhances cardiac repair | circulation. Circulation 136(Suppl_1). https://www.ahajournals.org/doi/abs/10.1161/circ.136.suppl_1.16195. Accessed 14 Mar 2019
Maeder ML, Gersbach CA (2016) Genome-editing technologies for gene and cell therapy. Mol Ther 24:430–446. https://doi.org/10.1038/mt.2016.10
McElreavey KD, Irvine AI, Ennis KT, McLean WH (1991) Isolation, culture and characterisation of fibroblast-like cells derived from the Wharton’s jelly portion of human umbilical cord. Biochem Soc Trans 19:29S
Meca-Cortés O, Guerra-Rebollo M, Garrido C et al (2017) CRISPR/Cas9-mediated Knockin application in cell therapy: a non-viral procedure for bystander treatment of glioma in mice. Mol Ther – Nucleic Acids 8:395–403. https://doi.org/10.1016/j.omtn.2017.07.012
Nemudryi AA, Valetdinova KR, Medvedev SP, Zakian SM (2014) TALEN and CRISPR/Cas genome editing systems: tools of discovery. Acta Nat 6:19–40
Reiser J, Zhang X-Y, Hemenway CS et al (2005) Potential of mesenchymal stem cells in gene therapy approaches for inherited and acquired diseases. Expert Opin Biol Ther 5:1571–1584. https://doi.org/10.1517/14712598.5.12.1571
Samsonraj RM, Rai B, Sathiyanathan P et al (2015) Establishing criteria for human mesenchymal stem cell potency. Stem Cells 33:1878–1891. https://doi.org/10.1002/stem.1982
Sebastiano V, Maeder ML, Angstman JF et al (2011) In situ genetic correction of the sickle cell anemia mutation in human induced pluripotent stem cells using engineered zinc finger nucleases. Stem Cells 29:1717–1726. https://doi.org/10.1002/stem.718
Shen Y, Zhang J, Yu T, Qi C (2018) Generation of PTEN knockout bone marrow mesenchymal stem cell lines by CRISPR/Cas9-mediated genome editing. Cytotechnology 70:783–791. https://doi.org/10.1007/s10616-017-0183-3
Sun S, Xiao J, Huo J et al (2018) Targeting ectodysplasin promotor by CRISPR/dCas9-effector effectively induces the reprogramming of human bone marrow-derived mesenchymal stem cells into sweat gland-like cells. Stem Cell Res Ther 9:8. https://doi.org/10.1186/s13287-017-0758-0
Técnico IS, Ircm CEA, Diderot UP (2015) Knockdown of monocyte chemotactic protein 1 in human mesenchymal stromal cells using two different approaches : shRNA and CRISPR-Cas 9
Ullah I, Subbarao RB, Rho GJ (2015) Human mesenchymal stem cells – current trends and future prospective. Biosci Rep. https://doi.org/10.1042/BSR20150025
van den Akker G, van Beuningen H, Blaney Davidson E, van der Kraan P (2016) CRISPR/CAS9 mediated genome engineering of human mesenchymal stem cells. Osteoarthr Cartil 24:S231. https://doi.org/10.1016/j.joca.2016.01.445
Vanoli F, Tomishima M, Feng W et al (2017) CRISPR-Cas9–guided oncogenic chromosomal translocations with conditional fusion protein expression in human mesenchymal cells. Proc Natl Acad Sci 114:3696–3701. https://doi.org/10.1073/pnas.1700622114
Xu X, Gao D, Wang P et al (2018) Efficient homology-directed gene editing by CRISPR/Cas9 in human stem and primary cells using tube electroporation. Sci Rep 8:11649. https://doi.org/10.1038/s41598-018-30227-w
Zhang Z, Zhang Y, Gao F et al (2017) CRISPR/Cas9 genome-editing system in human stem cells: current status and future prospects. Mol Ther – Nucleic Acids 9:230–241. https://doi.org/10.1016/j.omtn.2017.09.009
Zuk PA, Zhu M, Ashjian P et al (2002) Human adipose tissue is a source of multipotent stem cells. Mol Biol Cell 13:4279–4295. https://doi.org/10.1091/mbc.e02-02-0105
Author Contributions
A.G. conceptualized the outline and contents of the article. All authors contributed to writing and editing of the manuscript.
Conflict of Interest
The authors confirm that this article content has no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Golchin, A., Shams, F., Karami, F. (2019). Advancing Mesenchymal Stem Cell Therapy with CRISPR/Cas9 for Clinical Trial Studies. In: Turksen, K. (eds) Cell Biology and Translational Medicine, Volume 8. Advances in Experimental Medicine and Biology(), vol 1247. Springer, Cham. https://doi.org/10.1007/5584_2019_459
Download citation
DOI: https://doi.org/10.1007/5584_2019_459
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-45892-8
Online ISBN: 978-3-030-45893-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)