Abstract
Solid-state NMR spectroscopy proved to be a versatile tool for characterization of structure and dynamics of complex biochemical systems. In particular, magic angle spinning (MAS) solid-state NMR came to maturity for application towards structural elucidation of biological macromolecules. Current challenges in applying solid-state NMR as well as progress achieved recently will be discussed in the following chapter focusing on conceptual aspects important for structural elucidation of proteins.
To my father Werner Heise (1944–2013)
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Amyloid fibrils
- Biomolecular solid-state NMR spectroscopy
- Dynamic nuclear polarization
- Membrane proteins
1 Introduction
In the last decade, solid-state NMR spectroscopy has proven its enormous potential for structural investigations of biological macromolecules or macromolecular complexes. In contrast to X-ray crystallography, which requires crystalline samples, and solution-state NMR spectroscopy, which requires fast tumbling molecules and thus molecular weights of less than a few hundred kDa, solid-state NMR spectroscopy imposes no restrictions upon solubility, crystallizability, molecular size, or even purity of the sample. Solid-state NMR spectroscopy is a particularly powerful method for investigating large protein assemblies or aggregates, integral membrane proteins reconstituted into lipid bilayers, biominerals, protein–DNA complexes, or even full virus particles.
In solution-state NMR spectroscopy, rapid isotropic tumbling leads to a complete averaging of chemical shift anisotropy, homonuclear and heteronuclear dipolar couplings, and also, in the case of nuclei with spin >1/2, of quadrupolar interactions. In contrast, in solids the resonance frequency and the magnitude of internuclear couplings depend on the orientation of the molecule within the magnetic field. These anisotropic interactions can either be exploited for information on dynamics and orientation, or overcome. This can be accomplished in different ways:
-
If the molecules have no preferred orientation in the sample, signals are dominated by their chemical shift anisotropy or the quadrupolar interaction, which leads to a distinct powder pattern. Even in the absence of isotropic tumbling, however, molecules can rotate in a preferred direction resulting in a partial averaging of the powder pattern. Especially in lipid bilayers or liposomes, dynamics of membranes can be probed by the line shapes either in 31P spectra of the lipid head groups or of 2H spectra in selectively deuterated positions in lipid side chains.
-
Membrane proteins can be oriented macroscopically, e.g. by reconstitution into lipid bilayers on glass plates which are aligned perpendicular to the magnetic field. Since all membrane-reconstituted proteins adopt the same orientation in lipid bilayers, narrow lines are observed, and their chemical shifts as well as their dipolar couplings are a signature of the orientation [1].
-
Anisotropic interactions can be averaged out by magic angle spinning (MAS) NMR: [2] by rapidly spinning the sample around an axis inclined at an angle of 54.7° whereby dipolar couplings, chemical shift anisotropy, and first-order quadrupolar interactions are averaged to zero. In principle, line widths as small as in liquid-state NMR spectroscopy can be obtained (Fig. 1).
Fig. 1 Principle of magic angle spinning (MAS) solid state NMR. (a) Rotation of the sample rotor about the “magic angle” θ m = 54.7° with respect to the magnetic field B 0, i.e., the space diagonal. (b) Typical solid state NMR spectrum without MAS (left hand side, simulated data) and with MAS (right hand side). The arrow indicates the isotropic chemical shift δ iso
The rapid methodological development of solid-state NMR spectroscopy, in particular MAS NMR spectroscopy, during the last decade as well as more sophisticated labeling techniques for proteins have established the possibility of also applying multi-dimensional NMR spectroscopy to biological macromolecules and complex multi-component systems [3–5].
In the current chapter we focus on recent techniques and applications in MAS-NMR spectroscopy of proteins and biomolecules.
2 Sample Preparation
2.1 Isotope Labeling
Due to high sensitivity and resolution, 1H NMR detection is the basis of most solution-state NMR spectra of proteins. Solid-state 1H NMR spectra of proteins, however, are generally broadened beyond resolution due to spin relaxation in a network of dipolar coupled protons. Therefore, solid-state NMR spectroscopy of larger molecules like proteins critically depends on labeling with the stable isotopes 13C, 15N, and/or 2H. Depending on the method chosen for protein production, a variety of isotope labeling strategies has been developed [6–9].
For peptides below a length of about 40 amino acids, solid phase synthesis can be applied in which nascent peptides are bound to a bead and synthesized gradually in iterated cycles of deviating chemical environments. Thus, site-specific introduction of isotope labels at any desired position is possible.
Larger proteins are usually obtained by recombinant expression in E. coli, yeast [10], insect cells [11], or mammalian cells [12]. In these cases, the isotope labeling pattern depends critically on the carbon- and nitrogen-containing precursors in the growth medium. For 13C labeling, all intermediates of the glycolysis pathway and the citric acid cycle can serve as a carbon source. If the bacterial minimal growth medium contains glucose or its derivatives such as glycerol, pyruvate, or acetate as the sole carbon source and ammonium salts as the sole nitrogen source, uniformly 13C and 15N-labeled proteins can be obtained. Although such a uniform labeling pattern allows for collecting the maximum amount of spectroscopic information, the size of the protein as well as an unfortunate amino acid distribution might result in spectral overlap. In such cases, a reduction in the number of labeled amino acids might become crucial which can be achieved, e.g. by adding unlabeled amino acids to the growth medium. The success of this so-called reverse labeling depends on the metabolic pathway for the biosynthesis of the chosen amino acid [13, 14] which can be improved by the use of auxotrophic strains unable to synthesize the respective amino acids [15]. Complementary, type-selective amino acid labeling can be obtained by adding a labeled amino acid type to an otherwise unlabeled growth medium [16]. Atom position-specific labeling can be achieved by using reduced labeled precursors such as 1,3- or 2-13C-glycerol or 1- or 2-13C glucose resulting in characteristic 13C isotope distributions within each amino acid type [6, 9, 14, 17]. Incorporation of isolated 13C isotopes solely at methyl sites is obtained by using 3-13C-pyruvate as sole carbon source [18].
An even more sophisticated approach comprises protein synthesis by cell-free expression: as the amino acids added in a cell-free system are less exposed to bacterial metabolism, efficiency and selectivity of incorporation of specific isotope-labeled amino acids are only marginally hampered by isotope scrambling often observed in in vivo expression systems [19]. Even non-canonical amino acids can be incorporated allowing for site-specific labeling of proteins [20, 21].
At least for non-crystalline samples, 1H detection is still a major challenge. In solution NMR, the development of heteronuclear multidimensional NMR spectroscopy has paved the way for the assignment of backbone and side-chain resonances of proteins with molecular masses of over 30 kDa. This success has mainly been achieved through the production of highly deuterated samples because the replacement of protons with deuterons significantly improves the 1H resolution [22–25]. Innovative labeling schemes, e.g. with 5 % protonation at non-exchangeable sites, enable 1H-detection with high resolution resulting in determination of long range proton proton distances in solid-state MAS NMR spectroscopy [26]. However, protein deuteration is accompanied by several disadvantages. It not only depletes the number of 1H–1H distance restraints but also influences NMR resonance frequencies and cross polarization transfer efficiencies. In contrast, full protonation simplifies sample preparation and permits a more complete chemical shift assignment to be obtained from only one sample. Using the fully protonated 56-residue β1 immunoglobulin-binding domain of protein G (GB1) in a 1.6-mm rotor at 40 kHz MAS and 500 MHz proton frequency, proton line widths of 1 ppm and a sensitivity enhancement of up to four times compared to direct 13C and 15N detection could be observed. Apart from this, 3D pulse sequences transferring magnetization between heteronuclei such us CαNH, CONH, and NCαH even allowed full backbone and partial side-chain proton assignments [27].
A mixing of molecular species with different labeling patterns offers the possibility of obtaining intermolecular distance restraints [28].
Aiming at investigating a complex macromolecular surrounding such as whole cell preparations it has to be guaranteed that predominantly the protein of interest is isotope-labeled. In order to reduce the overall host protein content, single protein production, taking advantage of an mRNA interferase which cleaves all not genetically engineered mRNAs, constitutes a major milestone to a further improvement of sample quality [29].
2.2 Microcrystals and Macromolecular Complexes
The quality of solid-state NMR spectra critically depends on the properties of the sample. Depending on size, nature, and state of the protein, different ways of sample preparations can be used for solid-state NMR spectroscopy.
Pioneering solid-state NMR investigations on extensively isotope labeled proteins were performed on immobilized and often microcrystalline proteins. Especially for the later, well-resolved spectra could be obtained due to the absence of conformational disorder. Conformational ensembles of completely or partially unfolded proteins or multimers can be studied in frozen solution [30–32], where a lack of secondary structure does not result in an average random coil chemical shift but gives rise to a continuous distribution of chemical shifts [33]. Recently it was found that solid-state NMR spectroscopy can be performed on proteins >100 kDa even in solution due to transient sedimentation of large proteins at the rotor walls under fast MAS. Although these sediments are non-crystalline, the proteins are sufficiently immobilized suppressing motional averaging of dipolar interactions and chemical shifts [34, 35].
In contrast to solution-state NMR spectroscopy, the size of protein assemblies does not affect the line width in solid-state NMR spectroscopy. As only the number of inequivalent amino acids in a sample leads to spectral crowding and resonance overlap, large protein assemblies formed from one or only a few types of monomers are extremely suitable for solid-state NMR spectroscopy. Recent examples include a variety of amyloid fibrils [3, 36, 37], full virus particles [38, 39], and a secretion needle [40].
2.3 Amyloid Fibrils: Seeded Versus Spontaneous Fibrillation
A particular challenge associated with the study of assemblies of proteins in their non-native conformation is structural heterogeneity or polymorphism. As proteins may adopt different conformations upon aggregation, the structure of such fibrils can critically depend on the exact conditions of fibrillation. Minor variations of the pH, salt concentration [41], and even stirring of the solution [42] have been shown to have a tremendous influence on the morphology of the fibrils as well as the detailed molecular structure.
The fibrillation kinetics is characterized by a lag phase followed by a sigmoidal increase which is indicative of a nucleation-dependent aggregation, in which the formation of a nucleus is the rate-determining step. A lag phase can be circumvented by seeding with fibrils which have either been generated in vitro, or were isolated from in vivo material.
For amyloid fibrils of wild-type Aβ as well as a disease-related mutation thereof, sample homogeneity could be improved using a repeated seeding protocol [43, 44]. Increased homogeneity was ascribed to the selection of one fibril structure, which is kinetically as well as thermodynamically favored, out of a polymorphic mixture of different fibril types in the initial sample. However, the ultimate goal of this sort of study is the elucidation of disease-relevant conformations of the misfolded protein. As solid-state NMR studies rely on specific or uniform labeling with the NMR active isotopes 13C and/or 15N, structural investigations of amyloid fibrils purified from in vivo brain material – such as prion rods or fibrils generated from Alzheimer’s plaques or Lewy bodies – are still impossible. In such cases, templating of isotope-labeled recombinantly expressed proteins with brain-derived seeds becomes the method of choice for investigating fibrils in a conformation as close to the in vivo state as possible. First promising attempts include Aβ [45] or the yeast prion Ure2p [46].
2.4 Membrane Proteins in a Native-Like Environment
In terms of sample complexity, the preparation and investigation of membrane proteins is particularly challenging because of their intrinsic hydrophobic nature and the necessity of using a suitable membrane-mimicking environment (Fig. 2) [47]. For solid-state NMR the reconstitution into lipid bilayers is often the method of choice. Although incorporation of detergent-solubilized membrane proteins into liposomes occurs spontaneously by decreasing the detergent concentration, it has to be demonstrated that reconstituted membrane proteins are fully functional under physiological conditions of pH, salinity, and the presence of endogenous ligands. As an advantageous model of biological membranes, so-called bicelles became popular during the last decade due to their capability to align in a magnetic field. They are composed of a mixture of long chain (14–18 carbon atoms) and short chain (6–8 carbon atoms) surfactants and hence represent an intermediate between lipid vesicles and micelles. Since bicelles are not disrupted by MAS, isotropic solid-state 13C-, 15N-, and 31P-NMR spectroscopy can be applied for structural determination of molecules in membranes [48–51]. Finally, cellular membranes can be mimicked by discoidal nanoscale lipid bilayers, so-called nanodiscs, which are confined and stabilized by amphiphatic helical scaffold proteins [52]. They can be produced from a variety of lipids, are stable over a broad range of temperatures, and are accessible from both sides of the lipid bilayer. Since nanodiscs represent a more native environment than micelles or bicelles, they are an outstanding model system for understanding membrane protein function. Heteronuclear solid-state NMR studies of nanodisc-incorporated proteins demonstrated the utility of MAS NMR to study the structure of high molecular weight lipid protein complexes [53–55].
Schematic representation of most common membrane-mimicking environments compatible with solution and/or solid-state NMR studies of integral membrane proteins. Micelle (a), bicelle (b), nanolipoprotein particles also known as nanodiscs (c), and liposomes (d). The membrane protein is depicted in gray, detergents are colored in brown and lipids in blue. The apolipoprotein also known as membrane scaffold protein is represented as a green ring. Liposomes are scaled down by a factor of 2
3 Hardware Improvements: High Fields, Cryo-NMR, Fast MAS
One of the most obvious improvements during the last two decades of biomolecular NMR spectroscopy has been the development of high magnetic field strengths. As the chemical shift scales linearly with the external magnetic field, the spectral resolution increases linearly if the line widths are determined predominantly by the life time of the spin states (i.e. if the lines are homogeneously broadened as defined by Maricq and Waugh [56]). Furthermore, the signal intensity is proportional to the population difference between the two spin states of spin ½ nuclei given by the Boltzmann distribution. The higher the magnetic field, the higher the number of excitable spins in the low energy state resulting in an at least linear improvement of NMR-sensitivity as well. The strongest NMR instruments currently commercially available operate at a magnetic field strength of 23.5 T, corresponding to a proton Larmor frequency of 1 GHz.
The population difference given by the Boltzmann distribution can further be enhanced by lowering the sample temperature. Cryo-solid-state NMR at temperatures down to 90 K demonstrates that site-specific insights into molecular nature and dynamics of peptides and proteins can also be achieved in this temperature regime [57]. The increase in sensitivity, however, may be gained at the expense of spectral resolution which degrades substantially at temperatures below 210 K [58]. The observed line broadening reflects the freezing of molecular motions and side chain rotations into discrete populations. The dynamic transitions are reversible, at least in microcrystalline systems, excluding cold-denaturation or damage to the crystal lattice by freezing [57, 59, 60].
Recent developments in probe head technology have enabled MAS spinning rates of 70 kHz and more, which are particularly attractive in combination with direct proton detection [61, 62].
Finally, spin hyperpolarization techniques for obtaining spin polarizations exceeding the Boltzman limit by several orders of magnitude like optical polarization, para hydrogen induced hyperpolarization, or dynamic nuclear polarization (DNP) are subject of intense research. In the last section we will highlight recent break-throughs and latest contributions of DNP in the field of biomolecular solid-state NMR spectroscopy.
4 Basic Principles and Recent Pulse Sequence Improvements
4.1 Homogeneous Line Broadening and Proton Decoupling
Rapid exchange of energy quanta between nuclear spins in a 3D network by flip-flop transitions interferes with averaging by magic angle spinning, as the neighboring spins can change their spin state spontaneously and unpredictably during the rotor period. As a consequence rotational echoes decay fast and proton spectra remain homogeneously broadened even at ultra-high spinning speeds up to 70 kHz [56]. For high resolution, solid-state NMR spectroscopy of biological macromolecules depends on isotope labeling with 13C and 15N or on dilution of proton spins by extensive sample deuteration. Likewise, heteronuclear dipolar coupling of 13C or 15N spins to a network of strongly coupled proton spins prevents the refocusing of rare spins after each rotor period into a rotational echo. For high resolution in solid-state NMR spectroscopy, high power proton decoupling during evolution and detection periods is mandatory.
Homonuclear proton decoupling in the indirect evolution or in the direct detection period can be achieved by a net evolution of the protons around an effective field axis which is tilted by the magic angle of 54.7° from the magnetic field axis: in this interaction frame, the zero order term of the homonuclear dipolar Hamiltonian vanishes to zero. For indirect evolution periods this is achieved by irradiating with a strong rf field characterized by a resonance offset Ω that is matched to the rf field strength ω rf according to \( {\omega_{\mathrm{ rf}}}/\varOmega =\sqrt{2} \) [63]. Since the magnetization does not precess freely but nutates around a tilted axis, the chemical shift as well as heteronuclear dipolar couplings are reduced by a factor of \( 1/\sqrt{3} \). First-order correction terms of the average homonuclear dipolar Hamiltonian are averaged out by either switching the resonance offset between +Ω and −Ω (frequency switched Lee-Goldburg, FSLG) [64]. Alternating frequency offsets can also be mimicked by continuous phase modulation of the rf pulse (phase-modulated Lee-Goldburg, PMLG [65] or DUMBO [66]).
For homonuclear decoupling during acquisition, a net evolution of the magnetization around an axis tilted by the magic angle from the B 0 field can be achieved by toggling the interaction frame of the average Hamiltonian between x-, y-, and z-directions [67]. As long as MAS is slow compared to the toggling of the interaction frame, MAS and multi-pulse irradiation can be combined for averaging of anisotropic interactions (CRAMPS) [68]. For faster MAS rates above 30 kHz, where sample spinning may interfere with CRAMPS, homonuclear decoupling can also be achieved by applying phase-modulated spin lock fields alternating with time windows during which data points are acquired [69, 70].
For heteronuclear decoupling, the most straightforward decoupling technique is on-resonance rf irradiation of protons with an rf field of constant amplitude and phase (continuous wave decoupling, CW) [71] during evolution or detection periods. As CW decoupling leads only to the removal of zero order terms, better decoupling efficiencies can be obtained by phase modulations in the rf field [72–74]. However, as rf irradiation synchronized with the MAS rate may lead to interference of motion in real and spin space and thus to a recoupling of unwanted dipolar interactions instead of a decoupling, at high MAS rates extreme care has to be taken that the decoupling power is not in the same order of magnitude as the MAS rate. At ultrafast MAS rates it may be convenient to choose a decoupling power which is much lower than the MAS rate [62, 75].
4.2 Recoupling
Magic angle spinning narrows spectral lines and thus enhances resolution. On the other hand, valuable information about internuclear distances – encoded in anisotropic couplings – is lost. It is highly desirable to retrieve these interactions even in the presence of MAS for defined time intervals. Anisotropic interactions like homo- or heteronuclear dipolar couplings can be reintroduced selectively utilizing a second periodic modulation of the respective Hamiltonian which interferes with the averaging by MAS. This procedure is called recoupling (for recent reviews see [76, 77]). Homonuclear and heteronuclear dipolar recoupling can easily be incorporated into 2D experiments. Cross peaks between different spins are an indication of a dipolar coupling, i.e., spatial vicinity between nuclear spins.
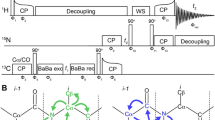
One of the most illustrative examples for recoupling of heteronuclear dipolar couplings is rotational echo double resonance (REDOR) [78]. In this recoupling scheme, the dipolar Hamiltonian is inverted twice per rotor period by applying 180°-pulses on one of the two rf channels (Fig. 3). This periodic inversion interferes with the periodic averaging of the dipolar coupling by MAS. As a consequence, a scaled dipolar Hamiltonian is retained for each spin pair which depends on the orientation of the two spins within the rotor. Whereas the classical REDOR-recoupling results in a distance-dependent dephasing of dipolarly coupled spins, rotational echo double resonance recoupling can also be utilized for coherence transfer (TEDOR) [79]. Recent improvements of this technique allow for selective recoupling of defined spin pairs circumventing dipolar truncation by means of selective pulses or z-filtering [80].
In REDOR-recoupling, two phase-alternating 180°rf-pulses every rotor period τ rot on the heteronuclear S-channel average out the effect of magic angle spinning on heteronuclear dipolar couplings. The 180°-pulse in the middle of the recoupling block on the I-channel refocuses the chemical shift Hamiltonian
The technique applied most frequently for heteronuclear dipolar recoupling is cross polarization. Heteronuclear dipolar interactions can facilitate polarization transfer between spins in the doubly rotating frame if two rf spinlock fields with appropriate field strengths are applied simultaneously on both channels. The ratio between the rf field amplitudes B 1 H and B 1 X on the proton and the heteronuclear rf channel under MAS at a spinning frequency ν rot has to match the Hartmann–Hahn condition:
The polarization transfer from protons to low-gamma nuclei leads to a polarization enhancement of γ H/γ X. Therefore, in almost all experiments, cross polarization from protons to low-gamma nuclei is used for excitation instead of a 90° pulse on the low-gamma nucleus itself. Further, long T 1 relaxation times of low-gamma nuclei, may be circumvented by initial cross polarization from protons.
Cross polarization can also be used for magnetization transfer between low-gamma nuclei like 15N and 13C as a mixing sequence in a multidimensional NMR experiment. Due to the substantial shift separation of about 120 ppm between carbonyl and Cα carbon atoms in proteins, the CP transfer can be tailored specifically to NCα- or NCO-transfer (SPECIFIC-CP) [81]. In protein NMR spectroscopy such a heteronuclear correlation experiment may be incorporated into multidimensional experiments and thus be used to residue-specific resonance assignment in NMR spectroscopy of immobilized peptides or proteins [82].
Likewise, homonuclear couplings can be reintroduced by interfering with the magic angle spinning: the simplest recoupling scheme is rotational resonance recoupling which occurs if the rotor frequency is equal to the chemical shift difference of two dipolarly coupled spins, a phenomenon which was discovered as early as 1966 by Andrew [83] and subsequently explained and exploited for distance measurements in the solid state by Levitt, Griffin and coworkers [84].
Applying a spin-lock pulse with an rf amplitude equal to one or two times the rotor frequency leads to a recoupling of heteronuclear dipolar interactions (rotary resonance recoupling) [85]; a spin-lock pulse with an amplitude half of the rotor frequency recouples homonuclear dipolar couplings (homonuclear rotary resonance HORROR) [86].
Homonuclear dipolar coupling can also be recoupled by a variety of rotor-synchronized symmetry-adapted pulse schemes developed by Levitt [87]. In these sequences, the rf amplitude is equal to a fixed ratio of the rotor frequency, and the phase of the rf field is incremented or alternated at a defined rate by phase increments specified by the symmetry of the recoupling scheme.
Broadband homonuclear recoupling schemes which lead in zeroth order approximation to a dipolar Hamiltonian containing single- or double-quantum two-spin operators, however, suffer from dipolar truncation [88–90]: in multi-spin systems, small dipolar couplings between distant spin pairs are attenuated by a strong dipolar coupling to a third (closer) spin. Thus, those recoupling sequences are not suitable for obtaining multiple long-range distance constraints in extensively isotope labeled biomolecules. One remedy against dipolar truncation is the band-selective recoupling of dipolar couplings, possibly in combination with sparse isotope labeling [91]. Alternatively, second-order recoupling sequences like proton-driven spin diffusion and variants thereof as well as proton-assisted second-order recoupling techniques described in the following paragraph have been discovered and developed for obtaining structural information in biomolecular solid-state NMR spectroscopy.
4.3 (Proton-Driven) Spin Diffusion and Second-Order Recoupling for Polarization Transfer
Spectral spin diffusion, i.e., the exchange of longitudinal magnetization between dipolarly coupled nuclear spins of roughly the same energy via flip-flop transitions, can also be utilized for magnetization transfer in homonuclear correlation experiments. Although the homonuclear dipolar Hamiltonian for an isolated spin pair is averaged to zero under MAS, dipolar couplings in a multi-spin system lead to non-zero higher order terms in the average Hamiltonian. Thus, spin diffusion between protons can be used for magnetization transfer in homonuclear 2D 1H-1H correlation experiments [92–94] or be detected indirectly on heteronuclei [95, 96]. Likewise, dipolar couplings between 13C or 15N spin pairs have non-zero higher order terms when coupled to an abundant proton spin network. As a consequence, proton-driven spin diffusion (PDSD) is affected by dipolar truncation to a much lesser degree than active zero-order recoupling schemes [97]. Thus, in principle, structure determination based solely on distance constraints obtained by PDSD is possible [98, 99]. In order to facilitate proton-driven spin diffusion at high MAS spinning speeds, heteronuclear dipolar couplings to protons can be actively recoupled, as exploited in the dipolar assisted rotational resonance (DARR) mixing sequence [100–102]. For fast MAS rates, heteronuclear recoupling to protons can be made more robust by varying the phase angle of proton irradiation using the phase-alternated recoupling irradiation scheme (PARIS) [103].
At fast MAS rates, the energy mismatch of a zero-quantum transition due to a large chemical shift difference between both carbon nuclei can be compensated for by irradiating the proton spins with an rf field strength equal to the sum of the spinning speed and the chemical shift difference. This mixed rotational and rotary resonance recoupling was termed MIRROR spin diffusion [104]. Finally, the proton-assisted recoupling (PAR) [105] scheme, which involves irradiation on both proton and carbon channels avoiding all rotary resonance and Hartmann-Hahn conditions, is characterized by a three-spin effective Hamiltonian and is thus also relatively robust with respect to dipolar truncation. A heteronuclear three spin order variant, which involves three different types of nuclei by irradiating three rf channels, is called proton-assisted insensitive nuclei cross polarization (PAIN-CP) [106]. This recoupling scheme may lead to valuable long-range N–C distance information in extensively isotope labeled samples.
4.4 Dynamics
Solid-state NMR spectroscopy techniques traditionally rely on dipolar couplings between nuclei for coherence transfer. However, proteins in lipid bilayers or in proteinaceous aggregates may exhibit residual large-scale or local motions which may lead to a modulation of the anisotropic interactions on the respective time scale: submicrosecond motions lead to an averaging of the dipolar couplings and thus to a line-narrowing, whereas motions in the millisecond regime may be detected in 2D exchange experiments. For motions between these time scales, the time-dependent modulation interferes with the NMR time scale and thus leads to a line-broadening which is not averaged out by MAS. Highly flexible protein regions may selectively be studied by classical solution NMR techniques where the INEPT (Insensitive nuclei enhanced by polarization transfer) [107] based coherence transfer without proton decoupling leads to selective excitation of mobile regions [108–111]. Anisotropic motions such as sidechain rotations as manifested in the order parameter, can be obtained from recoupling of dipolar interactions and chemical shifts (DIPSHIFT) [112].
On the other hand, exchange broadening by motions in an intermediate dynamic regime may lead to complete disappearance of signals, which may become visible at lower temperatures [113, 114].
5 Applications
5.1 Amyloid Fibrils
A field of research, on which solid-state NMR spectroscopy had a tremendous impact during the past decade, is the structural study of amyloid fibrils. Solid-state NMR spectroscopy can provide information on several aspects of the cross-β core structure of amyloid fibrils such as the localization of β-strands within the amino acid sequence and their relative arrangement within protofilaments and at the protofilament interface. Even high-resolution structures for the fibril core have been determined. An overview over emerging central motifs for amyloid structure is given in Fig. 4.
Structural motifs underlying amyloid formation: (a) antiparallel β-sheet, (b) parallel β-sheet, (c, d) parallel β-sandwich consisting of two β-sheets connected by a 180° turn as suggested for Aβ(1–40), (e, f) superpleated parallel cross-β-structure with more than two β-sheets as suggested for Ure2p and Sup35, (g) parallel β-helix consisting of two β-sheets with one monomer involved in two layers. In case of gray arrows, a cross section of the fibril axis is shown, whereas colored arrows indicate a side view parallel to the fibril axis. Every monomer is displayed in a different color
Solid-state NMR spectroscopy can furthermore be employed to identify alternative non-cross-β core structures and to characterize the fibril periphery. It also reports on dynamic processes in amyloid fibrils permitting, e.g. the differentiation between segments of static and dynamic disorder.
In the following we give a brief overview of selected amyloid systems, whose structural characterization has greatly benefited from the use of solid-state NMR spectroscopy. For a more detailed insight we refer to recent review articles [3, 36, 37, 115].
5.1.1 Aβ Peptide
The β-amyloid peptides Aβ(1–40) and Aβ(1–42) are the major constituents of senile plaques and cerebrovascular amyloid deposits found in the brains of patients with Alzheimer’s disease (AD) and cerebral amyloid angiopathy [116]. Aβ is an alternative cleavage product of the amyloid precursor protein (APP), a membrane protein of still unknown function [117]. Fibrils of Aβ and smaller peptides thereof have been extensively studied by solid-state NMR spectroscopy. Although a β-sandwich always seems to be the underlying motif, fibril structure details are not only highly susceptible to polymorphism but also depend critically on preparation conditions. Tycko and coworkers identified two different polymorphic forms of Aβ(1–40) fibrils, characterized by different chemical shifts and different fibril morphologies. These two conformations are obtained under different fibrillation conditions and can be transferred to the next fibril generation by seeding [42]. Two different supramolecular arrangements were suggested for these polymorphs [43].
A distinct type of polymorphism was observed in the Aβ(1-40)-Iowa mutant D23N [44, 118]. In the same fibril preparation both parallel and antiparallel β-structures coexisted, the latter representing the major conformation. This finding illustrates that a single disease-related mutation can have extensive consequences for amyloid structure.
As an approach to study the structure of Alzheimer’s disease-related Aβ fibrils, Aβ(1-40) fibril growth was directed by seeding with fibrils extracted from brain tissue of deceased AD patients. The resulting NMR data supported a β-strand-turn-β-strand motif similar to de novo-generated Aβ-fibrils. The chemical shifts, however, were significantly different from those observed for unseeded fibrils, suggesting structural differences between brain-derived and synthetic Aβ fibrils [45]. Recently, Reif et al. reported another fibril type composed of asymmetric dimers [119], whereas Bertini, Mao et al. further extended the structural diversity in the U-shaped β-strand-turn-β-strand motif of Aβ-fibrils, pointing towards a complex picture of Aβ-fibrillation [120].
Solid-state NMR studies of Aβ oligomers or protofibrils are extremely challenging due to their metastability. However, NMR data obtained for Aβ(1–40) [121] and Aβ(1–42) [122] oligomers demonstrate chemical shifts similar to the fibrillar peptides. Aβ(1–40) protofibrils could be stabilized by an antibody-derived fusion protein [123, 124]. The corresponding protofibril chemical shifts were indicative of a β-sheet structure in the same sequence region as for mature Aβ fibrils. However, the protofibril β-strands were less extended, and the analysis of chemical shifts suggested a closer relation to oligomers than to mature fibrils.
5.1.2 α-Synuclein
Amyloid fibrils of α-synuclein are the main component of Lewy bodies which are the pathological hallmark of Parkinson’s disease [125]. The physiological function of the 140-residue α-synuclein is still elusive. Three point mutations associated with early onset familial Parkinson’s disease have been identified so far [126–128]. Fibrillar α-synuclein was the first amyloid from a full-length protein studied by solid-state NMR spectroscopy [110, 129] and the spectral quality has subsequently encouraged further studies [130–134]. The protein is capable of forming a high variety of polymorphic forms. In different studies at least four different fibril types have been obtained even under rather similar fibrillation conditions. These deviating fibril types are characterized by differences in chemical shifts of identical residues as well as the exact location of well-ordered β-strands and of statically disordered parts in the rigid N-terminus. Unambiguous assignments for the first 30 N-terminal residues could only be obtained in one case [134]. In contrast, the mostly negatively charged 40 C-terminal residues were always found to be flexible, lacking a defined secondary structure. Analyzing NC-transfer or CC-transfer spectra of differentially isotope labeled fibril units, residues 40–90 were identified to form parallel in register β-sheets [3, 133] which are in agreement with EPR-studies [135].
The fibrillation kinetics of an A30P α-synuclein mutant was observed to be substantially slower, although chemical shifts were identical to one wild-type fibril form grown under identical conditions [136]. Studies on A53T α-synuclein fibrils revealed an extended β-sheet core [113] and slight perturbations of the chemical shifts, whereas for the E46K-mutation larger chemical shift deviations were observed [137].
Two artificial supertoxic mutants with one or three β-sheet breaking proline substituents, respectively, were designed based on the location of the β-sheets identified by solid-state NMR spectroscopy [32]. As expected, in vitro fibrillation as well as aggregation in HEK cells was retarded in both mutants, especially in the triple mutant, shifting the equilibrium towards oligomeric intermediates. Corresponding solid-state NMR spectra revealed a decreased rigid β-sheet core for the single mutant. For the triple mutant, a high degree of disorder was determined as indicated by broad and featureless lines with secondary chemical shifts characteristic for β-sheets. A higher toxicity of these artificial mutants was confirmed by expression in Drosophila melanogaster, Caenorhabditis elegans, and cultured mammalian neurons.
5.1.3 Yeast Prions with Glutamine/Asparagine-Rich Prion Domains
Certain proteins in yeast cells or fungi can adopt alternative β-sheet-rich conformations leading to epigenetic variations of protein function and thus to different phenotypes [138]. Since the underlying metastable conformational changes can be transferred horizontally, these proteins are termed yeast prions. Saccharomyces cerevisiae prions such as Ure2p, Sup35, Rnq1p, and Mod5 consist of a functional globular domain and a prion domain which is rich in asparagine and glutamine residues [139, 140]. In the prion conformation, the prion domains adopt a parallel, in-register β-sheet structure [46, 141–144] stabilized by Q/N-polar zippers [145]. For Sup35p and Ure2p, the ability to form prions was demonstrated to be retained after shuffling of the prion domain sequence, probably due to the high content of Q/N-residues allowing for different patterns of forming parallel, in-register β-sheet structures [146, 147].
Yeast prions show a high degree of structural polymorphism which may be linked to the existence of different yeast prion strains. Polymorphism has been detected by solid-state NMR spectroscopy for full-length Sup35p as well as for a heptapeptide from Sup35p [144, 148, 149]. Yeast prion-seeded samples of Ure2p exhibited different solid-state NMR spectra than non-seeded samples [46]. Likewise, NMR-signals from the isolated Ure2p-prion domain differed from the full-length protein pointing to extensive interactions of the globular C-terminal domain with the prion domain [46, 150].
5.1.4 Functional Amyloid: The Yeast Prion HET-s
The prion protein HET-s from Podospora anserina is a functional prion involved in heterokaryon incompatibility and, so far, the only yeast prion protein for which the prion state leads to a gain of function instead of a loss of function. Solid-state NMR spectra of HET-s exhibit remarkably high resolution indicative of high local order and structural homogeneity. This permitted the determination of a high-resolution structure [151–153]. In contrast to all other amyloid fibrils studied so far, the HET-s prion domain forms a β-solenoid with two windings per monomer and a triangular hydrophobic core (Fig. 5). Studies on the full-length protein revealed that the globular domain is not well structured but can be considered as a molten globule [154].
Structure of HET-s (218–289) in its amyloid form as revealed by solid-state NMR spectroscopy. The depicted bundle of 20 conformers was deposited in the Protein Data Band with accession code 2RNM [152]
Recently, HET-s in the amyloid form was used as model fibrils for the study of Congo Red binding. The binding interface was determined by cross polarization from unlabeled Congo Red to fully deuterated HET-s fibrils [155]. Based on the complex structure, a non-congophilic HET-s mutant could be designed, providing a structural rationale for the different sensitivity of amyloid-binding dyes for amyloids of different proteins or different prion strains/variants.
5.2 Membrane Proteins
A second field in structural biology, where solid-state NMR has proven to be useful, is the study of membrane proteins. As lipid bilayers – the natural environment of membrane proteins – tend to form liposomes or vesicles with rotational correlation times too large for solution-state NMR spectroscopy, solid-state NMR spectroscopy has been demonstrated to be a viable method to gain insight into complex membrane protein systems. In the following section we give a brief overview over selected systems where solid-state NMR could provide useful information. For a more comprehensive overview we refer to recent review articles [4, 146, 156, 157].
5.2.1 Influenza Ion Channel M2
The 97-residue M2 protein of the influenza A virus forms a tetrameric proton channel which is activated at low pH values. It has been shown to play an important role in virus replication. After infection of the host cell by endocytosis, the low endosomal pH opens the channel. The subsequent acidification of the viral core triggers the dissociation of the virus and thus induces expression of the viral genes. This ion channel is a target for the antiviral drugs amantadine and rimantidine which inhibit proton conduction and thus the unpacking of the virus. The S31N point mutation in the most recent circulating virus strains results in resistance against these drugs. Consequently, structural information about the drug binding site is of prevalent interest.
As the single transmembrane helix of the M2 protein alone, M2(22–46), is capable of tetramerization and proton conduction, it constitutes an ideal target for structural studies. Two high-resolution structures of a detergent-stabilized transmembrane helix construct bound to amantadine and of a construct containing the transmembrane helix and an adjacent amphipathic helix bound to rimantidine have been determined by X-ray crystallography [147] and solution NMR spectroscopy [158], respectively. However, both studies revealed contradicting drug binding sites, indicating that structure and function are sensitive to the lipid environment. An intense solid state NMR spectroscopy investigation of a 25-residue construct of the transmembrane helix reconstituted into liposomes solved the question. Solid-phase synthesis of the short peptide construct allowed for site selective isotope labeling of residues of interest and thus accurate resonance assignments in spite of line widths larger than 1 ppm. Chemical shift mapping of selected residues upon amantadine binding revealed only slight chemical shift deviations for most residues of the channel [159] except for Ser31 [160] (Fig. 6). Subsequent 13C-2H REDOR measurements on a site- selectively 13C-labeled M2 transmembrane helix complexed with fully deuterated amantadine revealed one strong binding site at Ser31 and one weak binding site at the C-terminal membrane side [161]. The orientation of the drug could be inferred from 2H-lineshapes. Finally, studies on a larger construct, including the amphipathic helix, revealed that the strong binding site persists in the longer construct while the weak binding site is obstructed by the amphipathic helix in liposome samples [162]. A comparison of spectra from full-length M2 in native E. coli membranes, liposome-reconstituted full-length M2, and a liposome-reconstituted construct containing the transmembrane and the amphipathic helices yielded comparable chemical shifts for the three complexes, thus confirming structural similarity of the shorter constructs to the full-length protein [163].
(a) Side view showing the transmembrane part of tetrameric M2 (22–46) with amantadine (Amt) bound to the high-affinity luminal site, (b) top view showing the Ser 31 and Val 27 pore radii of the transmembrane part of M4 binding AMT. The orientation of Amt is slightly tilted from the channel axis. The time-averaged Amt orientation is parallel to the channel axis. Reprinted with permission from Macmillan Publishers Ltd: Nature [161], copyright (2012)
Other questions of interest comprise the mechanism of proton conduction, selectivity, and pH gating. One single His37 residue in the transmembrane helix is responsible for proton selective conduction at low pH values, whereas at high pH values four His side chains constrict the channel. Two models for proton conduction had been proposed. The so-called shutter model suggests that, at low pH, protonation of the His residues leads to pore opening by electrostatic repulsion of the four positively charged rings and formation of a water wire which conducts protons via the Grotthus mechanism. According to the shuttle model, His 37 actively shuttles the protons into the virion by subsequent protonation and deprotonation accompanied by conformational ring flip motions (see e.g. ref [164] and references therein for more details).
In a recent investigation, Hong and coworkers studied the transmembrane domain of M2 in liposomes at high and low pH [164]. At high pH, distinct resonances for two different tautomers of the neutral imidazole ring were resolved, and cross peaks between those resonances in the PDSD spectra indicated an edge-to-face π-stack of the four histidines which obstructs the channel. At low pH the protonated imidazolium rings were found to undergo microsecond ring flip reorientations (faster than the NMR time scale), as measured by the order parameters obtained from recoupled one-bond N–C and H–C dipolar couplings of the aromatic rings, thus confirming the shuttle model. Determination of HN and HC dipolar couplings at low temperature, i.e., when the reorientation of the rings was frozen revealed slightly enlarged bond lengths, indicative of hydrogen bond formation. Proton spin diffusion experiments identified water as the sole hydrogen bonding partner [165]. Finally, different polarization transfer rates from water to the protein in the open state, at low pH, in the closed state at high pH, as well as in the amantadine-blocked state revealed a larger water contact surface for the open channel and a reduced water contact surface for the blocked channel [166].
A recent study of a construct containing the transmembrane helix and the amphipathic helix of the S31N mutant [167], which was reversely isotope labeled for four hydrophobic residues, facilitated sequential resonance assignments for 23 residues. All secondary chemical shifts were found to be characteristic for α-helical secondary structure. Peak doubling of most resonances as well as cross peaks between resonance pairs in PDSD spectra indicates that the tetramer is a dimer of asymmetric dimers instead of a symmetric homotetramer. Upon addition of amantadine, no significant shift changes were observed, which is in agreement with the drugs resistance of this mutant.
5.2.2 Seven Transmembrane Helix Receptors
Seven transmembrane helices (7-TM) are a structural motif common to a large family of photo- and chemoreceptor proteins. Most prominent examples of 7-TM proteins include (bacterial) rhodopsins and G protein-coupled receptors (GPCRs).
5.2.2.1 Bacterial Rhodopsins
In archaea and bacteria a family of 7-TM proteins with retinal as a prosthetic group serves two distinct functions. First, a light-driven energy conversion, such as carried out by bacteriorhodopsin from the extreme halophile Halobacterium salinarum [168] or by proteorhodopsin [169], converts light energy into an ion gradient, building up an electric potential across the membrane. Second, phototactic light sensing, such as performed by sensory rhodopsin [170], activates a signal transduction chain via a bound transducer molecule.
Rhodopsins of both classes have been successfully investigated using NMR spectroscopy. While progress in solution-state NMR has enabled the de novo structure determination of the two 7-TM proteins sensory rhodopsin II [171] and proteorhodopsin [172] solubilized in their monomeric form in detergent micelles, the power of solid-state NMR lies in the investigation of 7-TM receptors in their native environment and oligomeric state. In this respect the sensory rhodopsin system offers a remarkable example for the potential of solid-state NMR. In its native oligomeric state, the 7-TM receptor sensory rhodopsin, upon light excitation triggers a signal transduction chain via a tightly bound transducer protein, which is homologous to the two-component system of eubacterial chemotaxis. So far, the native oligomeric state of the receptor and transducer complex has not been formed in detergent micelles [173], impeding the use of solution-state NMR. In addition, pronounced intermolecular crystal packing contacts are found in the X-ray structure in close proximity to the receptor-transduced binding interface [174]. Solid-state NMR could be used to characterize the heterodimeric receptor–transducer complex in a lipid bilayer setting [175]. In this native-like condition, solid-state NMR data identified a considerably larger binding interface than found in the crystal structure, contributing to a better understanding of phototactic signal transduction.
Further examples for the use of solid-state NMR to study 7-TM proteins include the light-activated proton pump bacteriorhodopsin which could be studied in detail in its native environment, the purple membrane. Valuable high-resolution insights into the structure of the functional core – in particular the configuration of the covalently bound retinal at different states in the photocycle – could be obtained [176–180].
More recently several other bacterial rhodopsins such as proteorhodopsin and Anabaena sensory rhodopsin (ASR) have been investigated with solid-state NMR. Remarkable spectral resolution and sensitivity resulting in nearly complete resonance assignments were obtained [181–184]. Based on these data, specific insights into, e.g., the effects of hydration water on molecular dynamics [185] or the structural properties of the inner core region of these 7-TM systems could be generated [186–188]. In addition, protein–protein interactions could be studied with paramagnetic relaxation measurements. Using this approach, reliable information about the oligomeric state as well as the quaternary structure of ASR in a lipid bilayer environment was obtained (Fig. 7) [189].
Trimer model of S26CR1 Anabaena sensory rhodopsin (ASR). The individual monomers are shown in different colors. R1 side chains are shown in blue. Side chains of residues experiencing large intermolecular paramagnetic relaxation enhancement (PRE) effects and therefore spatially close (<15 Å) to the nitroxide of a neighboring monomer are shown in magenta. Adapted with permission from [189]. Copyright (2012) American Chemical Society
Overall, these exciting results indicate that solid-state NMR spectroscopy will provide increasingly detailed information about structure, dynamics, and function of rhodopsins in a near-native setting within the next few years.
5.2.2.2 G protein-Coupled Receptors
Acting as a central interface between external stimulus and cellular response, G protein-coupled receptors (GPCRs) control the majority of intercellular signaling cascades in humans. GPCRs play an important role not only in a large number of signaling cascades but also in very diverse physiological processes such as vision and smell as well as the regulation of blood pressure, body weight, and cell death. Due to their essential function in human physiology, it is not surprising that between 30% and 50% of all modern drugs are estimated to target GPCRs [190–192]. Although sharing the same topology of seven transmembrane helices with the class of (bacterial) rhodopsins, the investigation of GPCR structure is considerably more challenging, predominantly due to difficulties in sample preparation.
However, tremendous efforts have been carried out to solve the 3D-structure of GPCRs. Recently a breakthrough in protein crystallization attempts led to publication of a growing number of GPCR crystal structures (see, e.g., [193–195] for recent reviews). To stabilize crystal growth, artificial cofactors and protein engineering, such as large modification of potentially important loop segments, turned out to be necessary. Hence, additional information obtained under more native-like conditions is very desirable to provide complimentary insights into receptor dynamics as well as ligand binding and G-protein interactions [196].
While a number of NMR studies of GPCRs and their ligands have been carried out (reviewed, e.g., in [146] and [197]), no well-resolved NMR spectrum of a solubilized GPCR could be obtained so far, which is related to low protein expression yields, high amounts of misfolded receptor, and lack of stability when incorporated into a non-native membrane mimetic. On the contrary, solid-state NMR was used to study functional GPCRs with high precision [198–202]. In addition to the use of a more stable lipid bilayer environment, the advantage of solid-state NMR for the investigation of GPCRs is the ability to study non-deuterated membrane proteins. This enables the use of eukaryotic expression systems such as human or insect cell lines which do not tolerate a high level of deuterated buffer. While relaxation effects impede high resolution solution-state NMR studies of solubilized GPCRs without a significant degree of deuterium labeling, solid-state NMR does not face this problem and hence allows the characterization of functional GPCRs expressed in eukaryotic systems. In particular, in combination with the structures obtained using X-ray crystallography, it can be anticipated that solid-state NMR will provide critical information about ligand sensing and signal transduction in GPCRs.
6 Sensitivity Enhancement By Dynamic Nuclear Polarization
6.1 Theoretical Background
The fact that electron spins have a gyromagnetic ratio about 660 times higher than proton spins is exploited in dynamic nuclear polarization (DNP).
As already proposed by Albert Overhauser [203] in 1953 and subsequently confirmed experimentally by Carver and Slichter [204, 205], electron spin polarization in solid metals can be transferred to nuclear spins if the unpaired conducting electrons are saturated by irradiation with the corresponding electron paramagnetic resonance (EPR) frequency. As a result, nuclear spin polarization may be enhanced by the factor γ e/γ n. Likewise, stable organic radicals can be used as polarizing agents. Depending on the EPR-line width and the strength of the external magnetic field (and thus the line width of the EPR transition) the polarization transfer between electrons and nuclear spins in solid samples can occur via the Solid-Effect (SE), the Cross-Effect (CE) or Thermal Mixing (TM).
SE is a two-spin process, which relies on the excitation of forbidden zero or double quantum electron-nuclear two-spin transitions. As these transitions are forbidden, their excitation requires high power microwave irradiation. The efficiency of the transfer scales with the inverse square of the nuclear Larmor frequency. Another prerequisite for a selective irradiation of the zero or double quantum transition using SE DNP is a line width of the corresponding EPR-transition smaller than twice the nuclear Larmor frequency (Fig. 8). Nevertheless, by enhancing the microwave power with a resonator, SE has been demonstrated to lead to a sensitivity gain of about factor 128 at a field of 5 T [207]. TM and CE constitute three-spin transitions involving two electron spins and one nuclear spin at high radical concentrations. They are effective if the line width of the homogeneously or heterogeneously broadened EPR-transition is larger than the nuclear Larmor frequency, such that a three-spin transition is energetically neutral (similar to proton spin diffusion) [208, 209]. While thermal mixing is most effective at low fields, where the field-dependent line broadening due to the g-factor anisotropy is small, the cross effect can facilitate efficient polarization transfer at high magnetic fields and low temperatures, especially if biradicals are used.
Experimental 1H DNP enhancement profiles for the SE and the CE mechanisms showing the positions of positive and negative enhancement and their dependence on the microwave irradiation frequency (or, rather the magnetic field for a given microwave frequency) as well as on the electron and nuclear Larmor frequencies ω0S and ω0I , respectively. (a) A typical SE enhancement profile obtained with 40 mM trityl. (b) A typical CE enhancement profile obtained with 10 mM TOTAPOL (20 mM electrons). The EPR spectrum of each radical is shown on top. The lines connecting the data points are to guide the eye. Reprinted with permission from [206]. Copyright 2012, American Institute of Physics
The recent success of DNP as a hyperpolarization technique in solid-state NMR spectroscopy is mainly due to the following developments [210].
For high-resolution NMR investigations high magnetic fields are needed. Consequently, high-power high-frequency microwave sources are necessary to saturate the broad EPR lines at these fields. This can be facilitated by gyrotrons, which are able to produce stable microwaves of frequencies of up to 263 GHz when operating at the fundamental frequencies. Higher microwave frequencies can be obtained by operating a gyrotron with a given magnetic field at the second harmonic. This results in frequency doubling and facilitates microwave frequencies of up to 526 GHz, corresponding to a magnetic field strength of 18.8 T or a proton Larmor frequency of 800 MHz [211–213]. As the spin lattice relaxation times of nuclear spins have to be sufficiently long to facilitate polarization transfer, low temperatures below 90 K are required. This condition can now also be met for applications with MAS.
Paramagnetic centers used as source for the polarization transfer need to be compatible with the polarization mechanism. For SE-DNP, which is effective for isolated spin centers, a polarizing agent characterized by a narrow EPR transition line is needed. For this purpose, derivatives of the trityl radical or 1,3-bisdiphenylene-2-phenylallyl (BDPA) (Fig. 9) have been demonstrated to be well suited [214]. If polarization is to be provided by external paramagnetic dopants, a three-spin transition by the CE transfer mechanism is the method of choice. Then at least two paramagnetic centers have to be strongly dipolarly coupled. This prerequisite is fulfilled by stable biradicals such as TOTAPOL [215–217], which consists of two closely linked TEMPO-radical units.
For recent reviews of DNP in solids at high fields we refer to the following review articles [210, 218]. In the following section we will give an overview about recent developments and applications of this emerging technique.
6.2 Applications of DNP to Biomolecular NMR Spectroscopy
The applicability of DNP to biomolecules in the solid state was first demonstrated using lysozyme in a frozen glycerol/water matrix doped with TEMPO monoradicals. In this study, helium gas cooled to 40 K served as bearing and drive gas for MAS [219]. Subsequently, hyperpolarization by DNP has successfully been applied to a virus particle [220], to multidimensional heteronuclear spectroscopy of protein microcrystals of amyloidogenic peptides [221], and to a membrane receptor trapped in different stages of the photocycle [176, 177, 222].
Intending to improve sample preparation and to explore different experimental conditions, it was then realized that one potential limitation to biological application may be the line width. Since low temperatures below 90 K are required for efficient polarization transfer, lines may be severely broadened, either homogeneously or inhomogeneously:
At low temperatures, especially mobile side chains in a protein can be frozen in multiple static conformations, resulting in inhomogeneous line broadening. Particularly in spectra of hydrated microcrystals, line broadenings of up to 3–4 ppm are observed upon cooling to below a temperature of 200 K [59, 60]. For extensively deuterated samples, however, DNP enhancements of one order of magnitude can still be obtained at temperatures around 180 K, maintaining a reasonable resolution of 2D-spectra which is significantly increased compared to 90 K spectra [223].
In addition to inhomogeneous line broadening, the presence of paramagnetic dopants may lead to shortened T 2 nuclear relaxation times and thus a homogeneous line broadening. When the polarizing agents, however, are spatially separated from the nuclei of interest, narrow lines can also be observed with DNP [224]. Thus, the spatial distribution of radicals within the sample as well as their concentration also influences the resolution of spectra [225].
Signal enhancement for more elaborate NMR sequences involving multiple CP-transfer steps depends critically on the nuclear relaxation rates in the rotating frame of all nuclei involved. As these relaxation rates can be drastically reduced in the presence of paramagnetic dopants, it is not surprising that signal enhancement does not increase monotonically with increasing radical concentrations, but reaches a maximum at rather moderate radical concentrations [225].
Initial DNP NMR experiments on a complex biochemical system, i.e. amyloid fibrils of the model peptide GNNQQNY, yielded encouraging results [226]. Addition of biradicals to the sample did not result in significant line broadening and chemical shifts were preserved at low temperatures. Since chemical shifts before and after freezing the sample to 100 K were also identical, low temperatures do not appear to change the fibril structure at a molecular level.
Subsequently, for amyloid fibrils of the SH3 domain of the 83-residue phosphatidylinositol-3-kinase, valuable structural information could be provided by MAS-DNP-NMR [227]. For fibrils grown from a mixture of exclusively 15N labeled monomers and monomers labeled by using [2-13C]glycerol, intermolecular distance constraints were obtained from TEDOR experiments. High field spectra recorded at room temperature by conventional NMR spectroscopy provided only 30 intermolecular cross peaks for 83 residues, an observation which was ascribed to an interference of protein dynamics with decoupling, recoupling and cross polarization. In particular, no cross peaks could be observed for aromatic residues because of their twofold ring flips. In contrast, a DNP enhanced TEDOR spectrum recorded at a temperature of about 100 K and a field strength of 400 MHz 1H frequency revealed many additional intermolecular 15N–13C cross peaks, especially in the aromatic region. Although these spectra suffered from a high number of cross peaks and a lower resolution, 20 additional distance constraints could be determined with certainty (Fig. 10).
Comparison between room temperature and DNP-enhanced, low temperature intermolecular correlation spectra. (a–c) 750 MHz intermolecular 15N–13C correlations in PI3-SH3 fibrils recorded at 300 K with 16 days of acquisition. (d–f) The identical spectral regions recorded at 100 K and 400 MHz with DNP enhancement in 32 h of signal averaging. (g) Illustration of the 23 interstrand contacts established from 13C–15N cross-peaks in the 750 MHz spectra acquired at 300 K in panels a–c. (h) The 52 interstrand contacts established from the 400 MHz DNP-enhanced spectra recorded at 100 K shown in panels (d–f). Reprinted with permission from [227]. Copyright (2012) American Chemical Society
A recent investigation of a full virion, the bacteriophage Pf1, also benefitted greatly from reduced mobility as well as signal enhancement by DNP [228]. Whereas only protein signals could be assigned in earlier conventional solid-state MAS NMR studies at room-temperature, in DNP-enhanced spectra DNA signals were also assignable. The chemical shifts of the desoxyribose 13C atoms were indicative of 2′-endo/gauche conformations and anti-glycosidic bond conformations, while the chemical shifts of the DNA bases were consistent with an unusual structure with little or no base pairing, but base stacking. Further, signal splittings suggest some minor variations in the environment due to different interactions of the two (non-paired) DNA strands of the virion with the coat protein. Selected protein/DNA contacts could be identified in the spectra.
DNP enhancement was also used to investigate 40 nmol of a 25-residue signal peptide bound to the lipid-reconstituted 600-residue protein translocation complex SecY translocon [229]. Although double quantum filtering was mandatory to suppress the large natural abundance SecY background, a decent 2D-spectrum of the peptide could be obtained within 20 h of measurement time. For three of four isotope labeled amino acids the spin system could be identified by a sequential walk. The corresponding secondary chemical shifts were indicative of an α-helical secondary structure of the peptide in its bound form.
DNP signal enhancement also facilitated the structural investigation of the Asian cobra neurotoxin II bound to the ligand-gated ion channel nicotinic acetylcholine receptor (nAChR) obtained from the electric organ of an electric ray [230, 231]. Despite the low concentration of the 15N- and 1,3-13C- or 2-13C-glycerol-labeled toxin, a decent 2D 13C-13C correlation spectrum could be recorded within 14 h. Interestingly, storage of the sample at −20 °C resulted in a reduction of radicals close to the membrane surface leading to a loss of polarization enhancement and a drastic resolution increase for amino acid residues close to the membrane surface.
The high sensitivity provided by DNP enhancement allowed for 3D-NCACX/NCOCX-based sequential walks for selectively labeled samples of the model membrane protein Mistic, even though it was studied in its native cellular membrane environment without prior purification and reconstitution. Corresponding secondary chemical shifts are in agreement with the solution-state NMR structure, indicating that the protein was well folded [232]. The integral membrane protein PagL was also studied in extracted cell walls as well as whole E. coli cells. A significant DNP enhancement allowed for the detection of resonances from the overexpressed PagL protein as well as from an endogenous membrane-associated lipoprotein, lipid molecules, and RNA bases, for which even inter-base pair cross correlations were observed [233].
In all previous examples, diamagnetic biomolecules were studied by solid-state NMR spectroscopy using the biradical TOTAPOL as an external polarization agent. However, about 10% of biological macromolecules possess a paramagnetic (metal) center or a functional group or cofactor which is present as a (meta)stable radical during the reaction cycle or electron transfer process. The possibility to exploit these endogenous radicals as polarization sources appears intriguing. A first pioneering study using an endogenous radical for DNP enhancement in biomolecules involved the small electron transport protein flavodoxin. It contains a single non-covalently bound flavine mononucleotide which cycles between the oxidized quinone and the reduced radical semiquinone form. Due to the intrinsically low radical concentration, the DNP polarization transfer was governed by the solid effect, which unlike the cross effect does not rely on strong dipolar couplings between electron spins.
After deuteration of the protein to 85%, a DNP enhancement factor of 15 was achieved, demonstrating the feasibility of SE DNP using endogenous radicals despite a rather low field strength of only 212 MHz [234]. SE DNP has also been demonstrated for model complexes of the high-spin transition metals Mn2+ (S = 5/2) and Gd3+ (S = 7/2). It was found that the DNP enhancement depends strongly on the line width of the central (−1/2 → +1/2) electron spin transition. For Mn2+, the enhancement factor was limited to about 2 due to the hyperfine coupling to the 55Mn nucleus splitting the corresponding EPR transition and thus the DNP enhancement profile into six lines. For Gd3+, however, a signal enhancement factor of 13 was obtained when the line broadening of the central EPR transition by second order zero field splitting was small [235]. These studies show that utilizing metal centers as polarization source for DNP enhancement may in principle be possible.
While this review could only highlight certain examples, it can be stated that the combination of the substantial improvements, in particular in terms of the available hardware, sample preparation techniques, and novel and improved concepts of data acquisition and NMR methodology, extended considerably the limits of systems that can be studied today. For the near future, it can be anticipated that exciting new applications will emerge and that increasingly detailed insights will be obtained in more and more challenging biological systems using modern solid-state NMR spectroscopy.
Abbreviations
- APP:
-
Amyloid precursor protein
- ASR:
-
Anabaena sensory rhodopsin
- CE:
-
Cross-effect
- CRAMPS:
-
Combined rotation and multiple pulse spectroscopy
- CW:
-
Continuous wave
- DARR:
-
Dipolar assisted rotational resonance
- DNP:
-
Dynamic nuclear polarization
- EPR:
-
Electron paramagnetic resonance
- FSLG:
-
Frequency-switched Lee-Goldburg
- GPCR:
-
G protein-coupled receptor
- HORROR:
-
Homonuclear rotary resonance
- INEPT:
-
Insensitive nuclei enhanced by polarization transfer
- MAS:
-
Magic angle spinning
- PDSD:
-
Proton-driven spin diffusion
- PMLG:
-
Phase-modulated Lee-Goldburg
- PRE:
-
Paramagnetic relaxation enhancement
- REDOR:
-
Rotational echo double resonance
- SE:
-
Solid effect
- TEDOR:
-
Transferred echo double resonance
- TM:
-
Transmembrane helix
References
Opella SJ, Marassi FM (2004) Chem Rev 104:3587
Andrew ER, Bradbury A, Eades RG (1958) Nature 182:1659
Heise H (2008) Chembiochem 9:179
McDermott A (2009) Annu Rev Biophys 38:385
Wylie BJ, Rienstra CM (2008) J Chem Phys 128:052207
Higman VA, Flinders J, Hiller M, Jehle S, Markovic S, Fiedler S, van Rossum BJ, Oschkinat H (2009) J Biomol NMR 44:245
Böckmann A (2008) Angew Chem Int Ed Engl 47:6110
Guo C, Geng C, Tugarinov V (2009) J Biomol NMR 44:167
Lundstrom P, Teilum K, Carstensen T, Bezsonova I, Wiesner S, Hansen DF, Religa TL, Akke M, Kay LE (2007) J Biomol NMR 38:199
Boer E, Steinborn G, Kunze G, Gellissen G (2007) Appl Microbiol Biotechnol 77:513
Madden DR, Safferling M (2007) Methods Mol Biol 363:39
Armstrong N, Sun Y, Chen GQ, Gouaux E (1998) Nature 395:913
Waugh DS (1996) J Biomol NMR 8:184
Hong M, Jakes K (1999) J Biomol NMR 14:71
Whittaker JW (2007) Methods Mol Biol 389:175
Blanco FJ, Hess S, Pannell LK, Rizzo NW, Tycko R (2001) J Mol Biol 313:845
LeMaster DM, Kushlan DM (1996) J Am Chem Soc 118:9255
Lee AL, Urbauer JL, Wand AJ (1997) J Biomol NMR 9:437
Kigawa T, Muto Y, Yokoyama S (1995) J Biomol NMR 6:129
Noren CJ, Anthonycahill SJ, Griffith MC, Schultz PG (1989) Science 244:182
Young TS, Schultz PG (2010) J Biol Chem 285:11039
Tugarinov V, Kanelis V, Kay LE (2006) Nat Protoc 1:749
Gardner KH, Kay LE (1998) Annu Rev Biophys Biomol Struct 27:357
Chevelkov V, Rehbein K, Diehl A, Reif B (2006) Angew Chem Int Ed Engl 45:3878
del Amo JM, Fink U, Reif B (2010) J Biomol NMR 48:203
Asami S, Schmieder P, Reif B (2010) J Am Chem Soc 132:15133
Zhou DH, Shah G, Cormos M, Mullen C, Sandoz D, Rienstra CM (2007) J Am Chem Soc 129: 11791
Etzkorn M, Böckmann A, Lange A, Baldus M (2004) J Am Chem Soc 126:14746
Mao L, Vaiphei ST, Shimazu T, Schneider WM, Tang Y, Mani R, Roth MJ, Montelione GT, Inouye M (2010) J Struct Funct Genomics 11:81
Havlin RH, Tycko R (2005) Proc Natl Acad Sci USA 102:3284
Heise H, Luca S, de Groot BL, Grubmüller H, Baldus M (2005) Biophys J 89:2113
Karpinar DP, Balija MB, Kugler S, Opazo F, Rezaei-Ghaleh N, Wender N, Kim HY, Taschenberger G, Falkenburger BH, Heise H, Kumar A, Riedel D, Fichtner L, Voigt A, Braus GH, Giller K, Becker S, Herzig A, Baldus M, Jackle H, Eimer S, Schulz JB, Griesinger C, Zweckstetter M (2009) EMBO J 28:3256
Sakellariou D, Brown SP, Lesage A, Hediger S, Bardet M, Meriles CA, Pines A, Emsley L (2003) J Am Chem Soc 125:4376
Mainz A, Jehle S, van Rossum BJ, Oschkinat H, Reif B (2009) J Am Chem Soc 131:15968
Bertini I, Luchinat C, Parigi G, Ravera E, Reif B, Turano P (2011) Proc Natl Acad Sci 108: 10396
Tycko R (2011) Annu Rev Phys Chem 62:279
Tycko R (2006) Q Rev Biophys 39:1
Goldbourt A, Gross BJ, Day LA, McDermott AE (2007) J Am Chem Soc 129:2338
Goldbourt A, Day LA, McDermott AE (2010) J Biol Chem 285:37051
Loquet A, Sgourakis NG, Gupta R, Giller K, Riedel D, Goosmann C, Griesinger C, Kolbe M, Baker D, Becker S, Lange A (2012) Nature 486:276
Hoyer W, Antony T, Cherny D, Heim G, Jovin TM, Subramaniam V (2002) J Mol Biol 322: 383
Petkova AT, Leapman RD, Guo Z, Yau W-M, Mattson MP, Tycko R (2005) Science 307:262
Paravastu AK, Leapman RD, Yau WM, Tycko R (2008) Proc Natl Acad Sci USA 105:18349
Qiang W, Yau WM, Tycko R (2011) J Am Chem Soc 133:4018
Paravastu AK, Qahwash I, Leapman RD, Meredith SC, Tycko R (2009) Proc Natl Acad Sci USA 106:7443
Kryndushkin DS, Wickner RB, Tycko R (2011) J Mol Biol 409:263
Cross TA, Sharma M, Yi M, Zhou HX (2011) Trends Biochem Sci 36:117
De Angelis AA, Opella SJ (2007) Nat Protoc 2:2332
Diller A, Loudet C, Aussenac F, Raffard G, Fournier S, Laguerre M, Grelard A, Opella SJ, Marassi FM, Dufourc EJ (2009) Biochimie 91:744
Cho HS, Dominick JL, Spence MM (2010) J Phys Chem B 114:9238
Grelard A, Loudet C, Diller A, Dufourc EJ (2010) Methods Mol Biol 654:341
Bayburt TH, Carlson JW, Sligar SG (1998) J Struct Biol 123:37
Li Y, Kijac AZ, Sligar SG, Rienstra CM (2006) Biophys J 91:3819
Kijac AZ, Li Y, Sligar SG, Rienstra CM (2007) Biochemistry 46:13696
Kijac A, Shih AY, Nieuwkoop AJ, Schulten K, Sligar SG, Rienstra CM (2010) Biochemistry 49:9190
Maricq MM, Waugh JS (1979) J Chem Phys 70:3300
Bajaj VS, van der Wel PC, Griffin RG (2009) J Am Chem Soc 131:118
Franks WT, Zhou DH, Wylie BJ, Money BG, Graesser DT, Frericks HL, Sahota G, Rienstra CM (2005) J Am Chem Soc 127:12291
Linden A, Franks W, Akbey Ü, Lange S, van Rossum B-J, Oschkinat H (2011) J Biomol NMR 51:283
Thurber KR, Tycko R (2008) J Magn Reson 195:179
Bertini I, Emsley L, Lelli M, Luchinat C, Mao J, Pintacuda G (2010) J Am Chem Soc 132: 5558
Laage S, Sachleben JR, Steuernagel S, Pierattelli R, Pintacuda G, Emsley L (2009) J Magn Reson 196:133
Lee M, Goldburg I (1965) Phys Rev 140:A1261
Dvinskikh SV, Zimmermann H, Maliniak A, Sandstrom D (2005) J Chem Phys 122:44512
Vinogradov E, Madhu PK, Vega S (1999) Chem Phys Lett 314:443
Sakellariou D, Lesage A, Hodgkinson P, Emsley L (2000) Chem Phys Lett 319:253
Haeberlen U, Waugh JS (1968) Phys Rev 175:453
Gerstein BC, Pembleton RG, Wilson RC, Ryan LM (1977) J Chem Phys 66:361
Vinogradov E, Madhu PK, Vega S (2002) Chem Phys Lett 354:193
Lesage A, Sakellariou D, Hediger S, Elena B, Charmont P, Steuernagel S, Emsley L (2003) J Magn Reson 163:105
Schaefer J, Stejskal EO (1976) J Am Chem Soc 98:1031
Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG (1995) J Chem Phys 103:6951
Detken A, Hardy EH, Ernst M, Meier BH (2002) Chem Phys Lett 356:298
Fung BM, Khitrin AK, Ermolaev K (2000) J Magn Reson 142:97
Ernst M, Samoson A, Meier BH (2001) Chem Phys Lett 348:293
Ladizhansky V (2009) Solid State Nucl Magn Reson 36:119
De Paëpe G (2012) Annu Rev Phys Chem 63:661
Gullion T, Schaefer J (1989) J Magn Reson 81:196
Hing AW, Vega S, Schaefer J (1992) J Magn Reson 96:205
Jaroniec CP, Tounge BA, Herzfeld J, Griffin RG (2001) J Am Chem Soc 123:3507
Baldus M, Petkova AT, Herzfeld J, Griffin RG (1998) Mol Phys 95:1197
Schütz A, Wasmer C, Habenstein B, Verel R, Greenwald J, Riek R, Böckmann A, Meier BH (2010) Chembiochem 11:1543
Andrew ER, Clough S, Farnell LF, Gledhill TD, Roberts I (1966) Phys Lett 21:505
Raleigh DP, Levitt MH, Griffin RG (1988) Chem Phys Lett 146:71
Levitt MH, Oas TG, Griffin RG (1988) Isr J Chem 28:271
Nielsen NC, Bildsoe H, Jakobsen HJ, Levitt MH (1994) J Chem Phys 101:1805
Levitt MH (2002) In: Harris RK (ed) Encyclopedia of nuclear magnetic resonance: supplementary volume. Wiley, Chichester, p 165
Hohwy M, Rienstra CM, Griffin RG (2002) J Chem Phys 117:4973
Bayro MJ, Huber M, Ramachandran R, Davenport TC, Meier BH, Ernst M, Griffin RG (2009) J Chem Phys 130:114506
Hohwy M, Rienstra CM, Jaroniec CP, Griffin RG (1999) J Chem Phys 110:7983
Bayro MJ, Maly T, Birkett NR, Dobson CM, Griffin RG (2009) Angew Chem Int Ed 48:5708
Caravatti P, Neuenschwander P, Ernst RR (1985) Macromolecules 18:119
Brus J, Petrickova H, Dybal J (2003) Solid State Nucl Magn Reson 23:183
Brown SP (2007) Prog Nucl Magn Reson Spectrosc 50:199
Zhang S, Meier BH, Ernst RR (1992) Phys Rev Lett 69:2149
Lange A, Luca S, Baldus M (2002) J Am Chem Soc 124:9704
Grommek A, Meier BH, Ernst M (2006) Chem Phys Lett 427:404
Castellani F, van Rossum B, Diehl A, Schubert M, Rehbein K, Oschkinat H (2002) Nature 420:98
Manolikas T, Herrmann T, Meier BH (2008) J Am Chem Soc 130:3959
Takegoshi K, Nakamura S, Terao T (2001) Chem Phys Lett 344:631
Takegoshi K, Nakamura S, Terao T (2003) J Chem Phys 118:2325
Morcombe CR, Gaponenko V, Byrd RA, Zilm KW (2004) J Am Chem Soc 126:7196
Weingarth M, Demco DE, Bodenhausen G, Tekely P (2009) Chem Phys Lett 469:342
Scholz I, Huber M, Manolikas T, Meier BH, Ernst M (2008) Chem Phys Lett 460:278
De Paepe G, Lewandowski JR, Loquet A, Bockmann A, Griffin RG (2008) J Chem Phys 129: 245101
Lewandowski JR, De Paepe G, Griffin RG (2007) J Am Chem Soc 129:728
Morris GA, Freeman R (1979) J Am Chem Soc 101:760
Andronesi OC, Becker S, Seidel K, Heise H, Young HS, Baldus M (2005) J Am Chem Soc 127:12965
Siemer AB, Arnold AA, Ritter C, Westfeld T, Ernst M, Riek R, Meier BH (2006) J Am Chem Soc 128:13224
Heise H, Hoyer W, Becker S, Andronesi OC, Riedel D, Baldus M (2005) Proc Natl Acad Sci USA 102:15871
Helmus JJ, Surewicz K, Surewicz WK, Jaroniec CP (2010) J Am Chem Soc 132:2393
Munowitz MG, Griffin RG, Bodenhausen G, Huang TH (1981) J Am Chem Soc 103:2529
Heise H, Celej MS, Becker S, Riede D, Pelah A, Kumar A, Jovin TM, Baldus M (2008) J Mol Biol 380:444
Helmus JJ, Surewicz K, Nadaud PS, Surewicz WK, Jaroniec CP (2008) Proc Natl Acad Sci USA 105:6284
Böckmann A, Meier B (2010) Prion 4:72
Glenner GG, Wong CW (1984) Biochem Biophys Res Commun 120:885
Kang J, Lemaire H-G, Unterbeck A, Salbaum JM, Masters CL, Grzeschik K-H, Multhaup G, Beyreuther K, Muller-Hill B (1987) Nature 325:733
Tycko R, Sciarretta KL, Orgel JP, Meredith SC (2009) Biochemistry 48:6072
Lopez del Amo JM, Schmidt M, Fink U, Dasari M, Fändrich M, Reif B (2012) Angew Chem Int Ed 51:6136
Bertini I, Gonnelli L, Luchinat C, Mao J, Nesi A (2011) J Am Chem Soc 133:16013
Chimon S, Shaibat MA, Jones CR, Calero DC, Aizezi B, Ishii Y (2007) Nat Struct Mol Biol 14:1157
Ahmed M, Davis J, Aucoin D, Sato T, Ahuja S, Aimoto S, Elliott JI, Van Nostrand WE, Smith SO (2010) Nat Struct Mol Biol 17:561
Scheidt HA, Morgado I, Rothemund S, Huster D, Fandrich M (2011) Angew Chem Int Ed Engl 50:2837
Scheidt HA, Morgado I, Huster D (2012) J Biol Chem 287:22822
Spillantini MG, Schmidt ML, Lee VMY, Trojanowski JQ, Jakes R, Goedert M (1997) Nature 388:839
Kruger R, Kuhn W, Muller T, Woitalla D, Graeber M, Kosel S, Przuntek H, Epplen JT, Schols L, Riess O (1998) Nat Genet 18:106
Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, Stenroos ES, Chandrasekharappa S, Athanassiadou A, Papapetropoulos T, Johnson WG, Lazzarini AM, Duvoisin RC, DiIorio G, Golbe LI, Nussbaum RL (1997) Science 276:2045
Zarranz JJ, Alegre J, Gomez-Esteban JC, Lezcano E, Ros R, Ampuero I, Vidal L, Hoenicka J, Rodriguez O, Atares B, Llorens V, Tortosa EG, del Ser T, Munoz DG, de Yebenes JG (2004) Ann Neurol 55:164
Kloepper KD, Woods WS, Winter KA, George JM, Rienstra CM (2006) Protein Expr Purif 48:112
Kloepper K, Zhou D, Li Y, Winter K, George J, Rienstra C (2007) J Biomol NMR 39:197
Kloepper KD, Hartman KL, Ladror DT, Rienstra CM (2007) J Phys Chem B 111:13353
Vilar M, Chou HT, Luhrs T, Maji SK, Riek-Loher D, Verel R, Manning G, Stahlberg H, Riek R (2008) Proc Natl Acad Sci USA 105:8637
Loquet A, Giller K, Becker S, Lange A (2010) J Am Chem Soc 132:15164
Gath J, Habenstein B, Bousset L, Melki R, Meier B, Böckmann A (2012) Biomol NMR Assign 6:51
Chen M, Margittai M, Chen J, Langen R (2007) J Biol Chem 282:24970
Lemkau LR, Comellas G, Kloepper KD, Woods WS, George JM, Rienstra CM (2012) J Biol Chem 287:11526
Comellas G, Lemkau LR, Nieuwkoop AJ, Kloepper KD, Ladror DT, Ebisu R, Woods WS, Lipton AS, George JM, Rienstra CM (2011) J Mol Biol 411:881
Saibil HR, Seybert A, Habermann A, Winkler J, Eltsov M, Perkovic M, Castaño-Diez D, Scheffer MP, Haselmann U, Chlanda P, Lindquist S, Tyedmers J, Frangakis AS (2012) Proc Natl Acad Sci USA 109:14906
Wickner RB (1994) Science 264:566
Suzuki G, Shimazu N, Tanaka M (2012) Science 336:355
Shewmaker F, Wickner RB, Tycko R (2006) Proc Natl Acad Sci USA 103:19754
Baxa U, Wickner RB, Steven AC, Anderson DE, Marekov LN, Yau WM, Tycko R (2007) Biochemistry 46:13149
Wickner RB, Dyda F, Tycko R (2008) Proc Natl Acad Sci USA 105:2403
Shewmaker F, Kryndushkin D, Chen B, Tycko R, Wickner RB (2009) Biochemistry 48:5074
Chan JCC, Oyler NA, Yau WM, Tycko R (2005) Biochemistry 44:10669
Goncalves JA, Ahuja S, Erfani S, Eilers M, Smith SO (2010) Prog Nucl Magn Reson Spectrosc 57:159
Stouffer AL, Acharya R, Salom D, Levine AS, Di Costanzo L, Soto CS, Tereshko V, Nanda V, Stayrook S, DeGrado WF (2008) Nature 451:596
van der Wel PCA, Lewandowski JR, Griffin RG (2007) J Am Chem Soc 129:5117
Lewandowski JR, van der Wel PCA, Rigney M, Grigorieff N, Griffin RG (2011) J Am Chem Soc 133:14686
Loquet A, Bousset L, Gardiennet C, Sourigues Y, Wasmer C, Habenstein B, Schütz A, Meier BH, Melki R, Böckmann A (2009) J Mol Biol 394:108
Ritter C, Maddelein ML, Siemer AB, Luhrs T, Ernst M, Meier BH, Saupe SJ, Riek R (2005) Nature 435:844
Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH (2008) Science 319: 1523
Van Melckebeke H, Wasmer C, Lange A, Ab E, Loquet A, Böckmann A, Meier BH (2010) J Am Chem Soc 132:13765
Wasmer C, Schütz A, Loquet A, Buhtz C, Greenwald J, Riek R, Böckmann A, Meier BH (2009) J Mol Biol 394:119
Schütz AK, Soragni A, Hornemann S, Aguzzi A, Ernst M, Böckmann A, Meier BH (2011) Angew Chem Int Ed 50:5956
Hong M, Zhang Y, Hu FH (2012) Annu Rev Phys Chem 63:1
Smith SO (2010) Annu Rev Biophys 39:309
Schnell JR, Chou JJ (2008) Nature 451:591
Cady SD, Hong M (2008) Proc Natl Acad Sci USA 105:1483
Cady SD, Mishanina TV, Hong MJ (2009) Mol Biol 385:1127
Cady SD, Schmidt-Rohr K, Wang J, Soto CS, DeGrado WF, Hong M (2010) Nature 463:689
Cady S, Wang T, Hong M (2011) J Am Chem Soc 133:11572
Miao Y, Qin H, Fu R, Sharma M, Can TV, Hung I, Luca S, Gor'kov PL, Brey WW, Cross TA (2012) Angew Chem Int Ed 51:8383
Hu FH, Luo WB, Hong M (2010) Science 330:505
Hong M, Fritzsching KJ, Williams JK (2012) J Am Chem Soc 134:14753
Luo W, Hong M (2010) J Am Chem Soc 132:2378
Andreas LB, Eddy MT, Chou JJ, Griffin RG (2012) J Am Chem Soc 134:7215
Oesterhelt D, Stoeckenius W (1971) Nature 233:149
Béjà O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, Jovanovich SB, Gates CM, Feldman RA, Spudich JL, Spudich EN, DeLong EF (2000) Science 289:1902
Gordeliy VI, Labahn J, Moukhametzianov R, Efremov R, Granzin J, Schlesinger R, Buldt G, Savopol T, Scheidig AJ, Klare JP, Engelhard M (2002) Nature 419:484
Gautier A, Mott HR, Bostock MJ, Kirkpatrick JP, Nietlispach D (2010) Nat Struct Mol Biol 17:768
Reckel S, Gottstein D, Stehle J, Löhr F, Verhoefen M-K, Takeda M, Silvers R, Kainosho M, Glaubitz C, Wachtveitl J, Bernhard F, Schwalbe H, Güntert P, Dötsch V (2011) Angew Chem Int Ed 50:11942
Klare JP, Bordignon E, Doebber M, Fitter J, Kriegsmann J, Chizhov I, Steinhoff HJ, Engelhard M (2006) J Mol Biol 356:1207
Moukhametzianov R, Klare JP, Efremov R, Baeken C, Goppner A, Labahn J, Engelhard M, Buldt G, Gordeliy VI (2006) Nature 440:115
Etzkorn M, Seidel K, Li L, Martell S, Geyer M, Engelhard M, Baldus M (2010) Structure 18:293
Bajaj VS, Mak-Jurkauskas ML, Belenky M, Herzfeld J, Griffin RG (2009) Proc Natl Acad Sci USA 106:9244
Mak-Jurkauskas ML, Bajaj VS, Hornstein MK, Belenky M, Griffin RG, Herzfeld J (2008) Proc Natl Acad Sci USA 105:883
Harbison GS, Smith SO, Pardoen JA, Winkel C, Lugtenburg J, Herzfeld J, Mathies R, Griffin RG (1984) Proc Natl Acad Sci USA 81:1706
Jaroniec CP, Lansing JC, Tounge BA, Belenky M, Herzfeld J, Griffin RG (2001) J Am Chem Soc 123:12929
Higman VA, Varga K, Aslimovska L, Judge PJ, Sperling LJ, Rienstra CM, Watts A (2011) Angew Chem Int Ed Engl 50(36):8432
Shi L, Ahmed MAM, Zhang W, Whited G, Brown LS, Ladizhansky V (2009) J Mol Biol 386:1078
Shi L, Kawamura I, Jung K-H, Brown LS, Ladizhansky V (2011) Angew Chem Int Ed 50:1302
Wang S, Shi L, Okitsu T, Wada A, Brown LS, Ladizhansky V (2012) Biomol NMR Assign
Pfleger N, Worner AC, Yang J, Shastri S, Hellmich UA, Aslimovska L, Maier MS, Glaubitz C (2009) Biochim Biophys Acta 1787:697
Yang J, Aslimovska L, Glaubitz C (2011) J Am Chem Soc 133:4874
Pfleger N, Lorch M, Woerner AC, Shastri S, Glaubitz C (2008) J Biomol NMR 40:15
Ward ME, Shi L, Lake E, Krishnamurthy S, Hutchins H, Brown LS, Ladizhansky VJ (2011) J Am Chem Soc 133:17434
Hempelmann F, Holper S, Verhoefen MK, Woerner AC, Kohler T, Fiedler SA, Pfleger N, Wachtveitl J, Glaubitz C (2011) J Am Chem Soc 133:4645
Wang S, Munro RA, Kim SY, Jung KH, Brown LS, Ladizhansky V (2012) J Am Chem Soc 134:16995
Hopkins AL, Groom CR (2002) Nat Rev Drug Discov 1:727
Jacoby E, Bouhelal R, Gerspacher M, Seuwen K (2006) ChemMedChem 1:761
Klabunde T, Hessler G (2002) Chembiochem 3:928
Rosenbaum DM, Rasmussen SG, Kobilka BK (2009) Nature 459:356
Zhao Q, Wu BL (2012) Acta Pharmacol Sin 33:324
Katritch V, Cherezov V, Stevens RC (2013) Annu Rev Pharmacol Toxicol 53:531
Kobilka B, Schertler GFX (2008) Trends Pharmacol Sci 29:79
Tapaneeyakorn S, Goddard AD, Oates J, Willis CL, Watts A (2011) Biochim Biophys Acta 1808:1462
Ahuja S, Hornak V, Yan ECY, Syrett N, Goncalves JA, Hirshfeld A, Ziliox M, Sakmar TP, Sheves M, Reeves PJ, Smith SO, Eilers M (2009) Nat Struct Mol Biol 16:168
Park SH, Prytulla S, De Angelis AA, Brown JM, Kiefer H, Opella SJ (2006) J Am Chem Soc 128:7402
Goncalves JA, South K, Ahuja S, Zaitseva E, Opefi CA, Eilers M, Vogel R, Reeves PJ, Smith SO (2010) Proc Natl Acad Sci USA 107:19861
Struts AV, Salgado GF, Martinez-Mayorga K, Brown MF (2011) Nat Struct Mol Biol 18:392
Mertz B, Struts AV, Feller SE, Brown MF (2012) Biochim Biophys Acta 1818:241
Overhauser AW (1953) Phys Rev 92:411
Carver TR, Slichter CP (1953) Phys Rev 92:212
Carver TR, Slichter CP (1956) Phys Rev 102:975
Hu KN, Debelouchina GT, Smith AA, Griffin RG (2011) J Chem Phys 134:125105
Smith AA, Corzilius B, Barnes AB, Maly T, Griffin RG (2012) J Chem Phys 136:015101
Wind RA, Duijvestijn MJ, Vanderlugt C, Manenschijn A, Vriend J (1985) Prog Nucl Magn Reson Spectrosc 17:33
Wollan DS (1976) Phys Rev B 13:3671
Maly T, Debelouchina GT, Bajaj VS, Hu KN, Joo CG, Mak-Jurkauskas ML, Sirigiri JR, van der Wel PCA, Herzfeld J, Temkin RJ, Griffin RG (2008) J Chem Phys 128:052211
Felch KL, Danly BG, Jory HR, Kreischer KE, Lawson W, Levush B, Temkin RJ (1999) Proc IEEE 87:752
Becerra LR, Gerfen GJ, Temkin RJ, Singel DJ, Griffin RG (1993) Phys Rev Lett 71:3561
Becerra LR, Gerfen GJ, Bellew BF, Bryant JA, Hall DA, Inati SJ, Weber RT, Un S, Prisner TF, McDermott AE, Fishbein KW, Kreischer KE, Temkin RJ, Singel DJ, Griffin RG (1995) J Magn Reson Ser A 117:28
Haze O, Corzilius B, Smith AA, Griffin RG, Swager TM (2012) J Am Chem Soc 134:14287
Hu KN, Yu HH, Swager TM, Griffin RG (2004) J Am Chem Soc 126:10844
Song CS, Hu KN, Joo CG, Swager TM, Griffin RG (2006) J Am Chem Soc 128:11385
Matsuki Y, Maly T, Ouari O, Karoui H, Le Moigne F, Rizzato E, Lyubenova S, Herzfeld J, Prisner T, Tordo P, Griffin RG (2009) Angew Chem Int Ed Engl 48:4996
Matsuki Y, Takahashi H, Ueda K, Idehara T, Ogawa I, Toda M, Akutsu H, Fujiwara T (2010) Phys Chem Chem Phys 12:5799
Hall DA, Maus DC, Gerfen GJ, Inati SJ, Becerra LR, Dahlquist FW, Griffin RG (1997) Science 276:930
Rosay M, Zeri AC, Astrof NS, Opella SJ, Herzfeld J, Griffin RG (2001) J Am Chem Soc 123:1010
vanderWel PCA, Hu KN, Lewandowski J, Griffin RG (2006) J Am Chem Soc 128:10840
Bajaj VS, Mak-Jurkauskas ML, Belenky M, Herzfeld J, Griffin RG (2010) J Magn Reson 202:9
Akbey U, Linden AH, Oschkinat H (2012) Appl Magn Reson 43:81
Barnes AB, Corzilius B, Mak-Jurkauskas ML, Andreas LB, Bajaj VS, Matsuki Y, Belenky ML, Lugtenburg J, Sirigiri JR, Temkin RJ, Herzfeld J, Griffin RG (2010) Phys Chem Chem Phys 12: 5861
Lange S, Linden AH, Akbey Ü, Trent Franks W, Loening NM, Rossum B-J, Oschkinat H (2012) J Magn Reson 216:209
Debelouchina GT, Bayro MJ, van der Wel PCA, Caporini MA, Barnes AB, Rosay M, Maas WE, Griffin RG (2010) Phys Chem Chem Phys 12:5911
Bayro MJ, Debelouchina GT, Eddy MT, Birkett NR, MacPhee CE, Rosay M, Maas WE, Dobson CM, Griffin RG (2011) J Am Chem Soc 133:13967
Sergeyev IV, Day LA, Goldbourt A, McDermott AE (2011) J Am Chem Soc 133:20208
Reggie L, Lopez JJ, Collinson I, Glaubitz C, Lorch M (2011) J Am Chem Soc 133:19084
Linden AH, Lange S, Franks WT, Akbey Ü, Specker E, van Rossum B-J, Oschkinat H (2011) J Am Chem Soc 133:19266
Krabben L, van Rossurn BJ, Jehle S, Bocharov E, Lyukmanova EN, Schulga AA, Arseniev A, Hucho F, Oschkinat H (2009) J Mol Biol 390:662
Jacso T, Franks WT, Rose H, Fink U, Broecker J, Keller S, Oschkinat H, Reif B (2012) Angew Chem Int Ed 51:432
Renault M, Pawsey S, Bos MP, Koers EJ, Nand D, Tommassen-van Boxtel R, Rosay M, Tommassen J, Maas WE, Baldus M (2012) Angew Chem Int Ed 51:2998
Maly T, Cui DT, Griffin RG, Miller AF (2012) J Phys Chem B 116:7055
Corzilius B, Smith AA, Barnes AB, Luchinat C, Bertini I, Griffin RG (2011) J Am Chem Soc 133:5648
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Müller, H., Etzkorn, M., Heise, H. (2013). Solid-State NMR Spectroscopy of Proteins. In: Heise, H., Matthews, S. (eds) Modern NMR Methodology. Topics in Current Chemistry, vol 335. Springer, Berlin, Heidelberg. https://doi.org/10.1007/128_2012_417
Download citation
DOI: https://doi.org/10.1007/128_2012_417
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-37990-1
Online ISBN: 978-3-642-37991-8
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)