Abstract
Mammalian expression systems such as Chinese hamster ovary (CHO), mouse myeloma (NS0), and human embryonic kidney (HEK) cells serve a critical role in the biotechnology industry as the production host of choice for recombinant protein therapeutics. Most of the recombinant biologics are glycoproteins that contain complex oligosaccharide or glycan attachments representing a principal component of product quality. Both N-glycans and O-glycans are present in these mammalian cells, but the engineering of N-linked glycosylation is of critical interest in industry and many efforts have been directed to improve this pathway. This is because altering the N-glycan composition can change the product quality of recombinant biotherapeutics in mammalian hosts. In addition, sialylation and fucosylation represent components of the glycosylation pathway that affect circulatory half-life and antibody-dependent cellular cytotoxicity, respectively. In this chapter, we first offer an overview of the glycosylation, sialylation, and fucosylation networks in mammalian cells, specifically CHO cells, which are extensively used in antibody production. Next, genetic engineering technologies used in CHO cells to modulate glycosylation pathways are described. We provide examples of their use in CHO cell engineering approaches to highlight these technologies further. Specifically, we describe efforts to overexpress glycosyltransferases and sialyltransfereases, and efforts to decrease sialidase cleavage and fucosylation. Finally, this chapter covers new strategies and future directions of CHO cell glycoengineering, such as the application of glycoproteomics, glycomics, and the integration of ‘omics’ approaches to identify, quantify, and characterize the glycosylated proteins in CHO cells.


Graphical Abstract
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Chinese hamster ovary
- CHO
- CRISPR/Cas9
- Fucosylation
- Glycoengineering
- Glycomics
- Glycoproteomics
- Mammalian expression systems
- N-linked glycosylation
- O-linked glycosylation
- Sialylation
- TALEN
- ZFN
1 Introduction
Both N-glycosylation and O-glycosylation pathways serve as key targets for mammalian cell engineering efforts. The efficiency and control of glycosylation in recombinant protein production is critical, because changes in protein glycosylation can affect circulatory half-life, bioactivity, and product quality. Improving the degree of glycosylation and sialylation can reduce clearance of the therapeutic product from the patient. Additionally, control of glycan antennarity helps to maintain lot-to-lot consistency during the drug production. Controlling fucosylation has important effects on therapeutic efficacy by regulating antibody-dependent cellular cytotoxicity (ADCC). Decreasing core fucosylation can improve antibody effector function and clinical efficacy. In summary, glycosylation control is crucial during the process of biotherapeutics development. This section introduces the Chinese hamster ovary (CHO) glycosylation pathway, as well as the importance of sialylation and fucosylation.
1.1 Glycosylation
Therapeutic glycoproteins include several classes, such as monoclonal antibodies (mAbs), immunoglobulin G fragment crystallizable domain (Fc)-fusion proteins (Fc-fusion proteins), enzymes, hormones, cytokines, growth factors, and hormones [1,2,3]. Overall, the biotechnology industry generates billions of dollars of sales from these glycoproteins [4]. The increasing demand for biotherapeutics for the treatment of cancer, autoimmune disorders, infectious diseases, genetic disorders, and metabolic disorders requires the development and precise control of glycotherapeutics production.
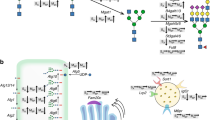
An overview of both N-glycosylation and O-glycosylation is shown in Fig. 1. During N-glycosylation, various carbohydrate chains are added to asparagine (Asn) [5] residues of proteins [5]. In contrast, O-glycosylation involves the addition of carbohydrate chains to serine (Ser) or threonine (Thr) [6]. While N-glycans are the most common modification in biotherapeutics such as mAbs, there are examples of therapeutic glycoproteins, such as erythropoietin (EPO) and etanercept, that also contain O-glycosylation [7]. Glycosylation is a critical post-translational modification found in most biotherapeutics; interestingly, the cellular process generates structural diversity that includes a number of different structures even for a single protein from one organism. The variety of glycoforms expands dramatically when the protein is produced in another host cell or species even under different reactor conditions. Most importantly, the pattern of glycosylation can play a major role in modulating a number of product quality characteristics [8].
Examples of N- and O-linked glycosylation. N-linked glycosylation involves the asparagine (Asn) [5] residue, whereas O-linked glycans extend from serine (Ser) or threonine (Thr) residues. GlcNAc: N-acetylglucosamine, GalNAc: N-acetylgalactosamine
A prerequisite for N-glycosylation is the requirement that N-glycans be linked to the Asn of the Asn-X-Ser/Thr consensus sequence, where X represents any amino acid except for proline [9]. A similar consensus sequence for O-linked glycosylation has not been identified [8]. As proteins are processed through the endoplasmic reticulum (ER) and Golgi apparatus prior to secretion, a number of enzymes can act to shorten or extend the N-glycan chain, as shown in Fig. 2. Since the enzymes do not act on every protein that traverses a particular compartment, the stochastic nature of the interactions creates heterogeneity, owing to the variability in glycosylation site occupancy and the diversity of glycoforms that are formed during passage through the secretory apparatus. In addition, there is continuous interplay between enzymes and oligosaccharide substrates. Since more than one enzyme can act on a glycan substrate, a wide arsenal of glycoproteins can be generated [10, 11].
Overview of N-linked glycosylation generating biantennary sialylated glycans. During N-linked glycosylation, various enzymes extend and trim the glycoprotein as it passes from the endoplasmic reticulum to the Golgi apparatus. Abbreviations: Glc I/II glucosidase I/II, ER Man I endoplasmic reticulum mannosidase I, Man I mannosidase I, GnT I N-acetylglucosaminyltransferase I, FucT C6 α(1,6)-fucosyltransferase, Man II mannosidase II, GnT II N-acetylglucosaminyltransferase II, β4GalT β-1,4-galactosyltransferase, SiaT sialyltransferase
The complex N-linked glycosylation reaction network shown in Fig. 2 involves glycosidases and glycosyltransferases that catalyze enzymatic modifications in different cellular compartments. First, the biosynthesis of mammalian N-glycans begins with the transfer of N-acetylglucosamine-1-phosphate (GlcNAc-P) from uridine diphosphate-N-acetylglucosamine (UDP-GlcNAc) to the dolichol phosphate (Dol-P) lipid carrier to generate dolichol pyrophosphate N-acetylglucosamine (Dol-P-P-GlcNAc) at the cytoplasmic face of the ER membrane [12]. Next, sugars are sequentially added to Dol-P-P-GlcNAc to form an oligosaccharide precursor known as Dol-P-P-GlcNAc2Man5 [12]. The structure is then flipped to the ER side and further extended to generate Glc3Man9GlcNAc2-P-P-Dol. Oligosaccharyltransferase (OST) identifies the consensus sequence (Asn-X-Ser/Thr) in the nascent polypeptide and transfers Glc3Man9GlcNAc2 from the dolichol-linked donor to the side chain amide of Asn, while releasing the Dol-P-P during the process [13]. The glucose residues on the precursor are then sequentially trimmed by ER alpha-glucosidase I and ER alpha-glucosidase II to form a monoglucosylated glycan. This intermediate plays a role in the ER lectin chaperones calnexin/calreticulin-associated glycoprotein folding control cycle [14]. When the precursor is folded, it is next trimmed to yield Man8GlcNAc2-protein before exiting the ER; this step is catalyzed by ER alpha-mannosidase I. The Man8GlcNAc2 glycoform is translocated into the cis-Golgi apparatus, where it is trimmed again, to Man5GlcNAc2, a key intermediate along the pathway to form hybrid and complex N-glycans, and sometimes found as a final glycan product; this step is catalyzed by Golgi alpha-mannosidases I.
In order to generate hybrid and complex N-glycans, N-acetylglucosaminyltransferase I (GnT-1 or Mgat1) is required to add GlcNAc to Man5GlcNAc2 in the medial Golgi apparatus [12]. N-Glycans are trimmed by Golgi alpha-mannosidase II, which removes two mannoses from GlcNAcMan5GlcNAc2 to generate GlcNAcMan3GlcNAc2. Hybrid N-glycans result from the incomplete removal of mannose residues, which occurs when a structure such as GlcNAcMan5GlcNAc2 undergoes no further extension or trimming and the structure ends up with one or two terminal Man residues. In an alternative process, GlcNAc can be added to the innermost Man group by the enzyme beta-1,4-N-acetylglucosaminyltransferase III (GnT-III or Mgat3) in the medial Golgi apparatus, a process which generates bisecting GlcNAc structures that alter the capacity for other downstream enzymes to act on the glycan structure.
The precursor for all multi-antennary complex N-glycans is GlcNAc2Man3GlcNAc2, which is generated by the action of beta-1,2-N-acetylglucosaminyltransferase II (GnT-II or Mgat2) that adds GlcNAc to the GlcNAcMan3GlcNAc2 structure. Tri-antennary and tetra-antennary branches are created through the addition of GlcNAc at the alpha-(1,3)-mannose site by N-acetylglucosaminyltransferase IV (GnT-IV or Mgat 4) and at the alpha-(1,6)-mannose site by N-acetylglucosaminyltransferase V (GnT-V or Mgat 5).
There can be further modifications, such as fucosylation, branch extension, and sialylation, which generate even more complex glycans. Fucosylation occurs in the trans Golgi apparatus with the addition of core alpha-(1,6)-fucose to the GlcNAc adjacent to Asn of the N-glycan by alpha-(1,6)-fucosyltransferase. Branch extension involves the addition of a beta-linked galactose residue to GlcNAc, which yields Gal-beta-1-4GlcNAc, also known as acetyl lactosamine (LacNAc). For sialylation, terminal Gal residues can be acted upon by alpha-(2,3)- or alpha-(2,6)-sialyltransferases that add sialic acid residues to the glycan [12].
One reason for the widespread use of CHO cell lines in biotechnology is their capacity to produce complex glycans that are compatible with the human immune system [1, 15]. Alternative mammalian cell lines can also produce biopharmaceuticals, but their use is not as widespread in industry because of their potential for immunogenicity and difficulty in manufacturing scale-up; examples include baby hamster kidney (BHK), murine myeloma and hybridoma cell lines (NS0 and Sp2/0), and human host cell lines, such as human embryonic kidney (HEK-293) and human retinal cells (PER.C6) [1, 2, 16].
When glycans are generated outside of human hosts, it is critical to avoid the production of non-human glycans, such as terminal Gal-alpha-1,3-Gal linkages (alpha-Gal) and N-glycolylneuraminic acid (Neu5Gc) residues, which may result in adverse immunogenic reactions if given to humans with a sensitivity to these residues [1, 17]. Mouse cells such as NS0 have an alpha-1,3-galactosyltransferase enzyme that produces glycans containing the alpha-Gal linkage [18]. The second potential immunogenic reaction from Neu5Gc is common in all non-primate mammalian cells, owing to the presence of the enzyme N-acetylneuraminic acid hydroxylase, which converts cytidine monophosphate (CMP)-N-acetylneuraminic acid (Neu5Ac) to CMP-Neu5Gc in all mammals other than old-world primates [1, 19]. Humans exhibit a circulating polyclonal anti-Neu5Gc antibody response, so it is desirable to avoid Neu5Gc in biotherapeutics production [1, 17]. In contrast to the alpha-Gal epitope, Neu5Gc can be metabolically incorporated into glycoforms during cell culture from metabolites in cell culture media. Mouse myeloma cells (NS0 and Sp2/0) thus exhibit the highest potential for immunogenicity because they express higher levels of alpha-Gal and Neu5Gc than CHO cells, which can be an issue if biotherapeutics with these modifications are provided to patients at large doses or for long periods [19,20,21]. These subtle differences in glycosylation processing are one of the principal reasons why CHO cells are preferred for bioproduction.
Aside from immunogenic epitopes, glycosylation patterns in CHO cells and humans often differ in other ways too [22]. One reason is that CHO cells lack bisecting GlcNAc residues because they typically do not express GnT-III; the resulting difference may affect the efficacy of the glycotherapeutics [23]. Human cells contain GnT III and can produce glycans with bisecting GlcNAc; in comparison, NS0 and SP2/0 cells are able to generate only a portion of glycans with bisecting GlcNAc residues [24].
Overall, the glycoform profiles on glycoproteins can vary widely depending on the cell lines, growth, and bioreactor conditions such as pH, temperature, media, and feeding strategies. The interplay of various glycosylation enzymes is responsible for the great diversity of glycoproteins. Some examples relevant to glycotherapeutics are shown in Fig. 3. Specifically, the degree of antennarity varies across glycoproteins. Glycosylation in biotherapeutics directly affects product quality because it plays a role in solubility, stability, protease resistance [25], aggregation [1, 2], serum half-life [26], immunogenicity [8], efficacy [27, 28], and ligand binding [29].
These impacts of glycosylation highlight the need for glycoengineering in order to yield glycotherapeutics with consistent and desirable glycoform profiles. In the next section of the chapter, we examine targets and review genetic engineering approaches to control glycosylation, such as increasing the expression of glycosylation and sialylation enzymes, or reducing the expression of sialidase cleavage and fucosylation enzymes. Recently, multiple genes have been modified simultaneously, and new strategies such as zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein-9 (Cas9) offer new technologies for glycoengineering applications.
1.2 Sialylation
Sialic acid addition is the final step of the N-glycosylation pathway. While both Neu5Ac and Neu5Gc are present in CHO cells, humans lack Neu5Gc. Sialic acid is a negatively charged acidic 9-carbon sugar moiety that is typically attached by an alpha-glycosidic linkage on the C3- or C6-hydroxyl group of terminal galactose by alpha-2,3-sialyltransferases (ST3) or alpha-1,6-sialyltransferases (ST6) individually [30,31,32,33]. The sialic acid moiety may also on occasion be attached to the C8 position of sialic acid to form sialic acid multimers, but this is not typically observed for glycoprotein therapeutics [34]. As the terminal end cap, sialic acid is especially relevant for the half-life and bioactivity of recombinant glycoproteins. The removal of sialic acid by sialidase exposes the terminal galactose, and its cleavage often decreases circulatory half-life. Without sialic acid capping the glycoprotein, the galactose molecule is recognized by the hepatocyte asialoglycoprotein receptor and is cleared from the body [32, 35, 36]. For improving the circulatory half-life of recombinant therapeutics, preventing recognition by this receptor is desirable. Additionally, fully sialylated glycoproteins can increase the size and overall charge of the molecule. Therefore, it is often desirable to enhance or maximize sialylation in CHO cells to improve the production of recombinant therapeutics [14]. Of course, for the case of biosimilars, it may also be relevant to match the sialylation profile of the innovator molecule. If the host cell line of the innovator and biosimilar are different from each other, matching the sialylation profile can be even more difficult. Therefore, both sialyltransferases and sialidases are targets for genetic engineering, because they affect opposing processes.
1.3 Fucosylation
Fucosylation, or the addition of fucose to glycoforms, occurs through both de-novo and salvage pathways. In the first pathway, d-glucose uptake into the cytoplasm generates guanosine diphosphate (GDP)-mannose. The enzymes GDP-mannose 4,6-dehydratase and GDP-keto-6-deoxymannose 3,5-epimerase, 4-reductase convert GDP-mannose into GDP-fucose [37]. In contrast, the salvage pathway utilizes l-fucose from extracellular and lysosomal sources. Fucokinase phosphorylates l-fucose into l-fucose-1-phosphate, and GDP-fucose pyrophosphorylase (GFPP) converts l-fucose-1-phosphate to GDP-fucose. GDP-fucose is subsequently transported to the Golgi apparatus by the GDP-fucose transporter and fucose is added onto the glycan chains of maturing glycoproteins by fucosyltransferases. Thus, enzymes are important in fucosylation and they are also important targets for cell engineering.
This step of glycosylation is critically important for antibody fragment crystallizable (Fc) receptor-mediated activity, which can strongly affect ADCC activity. During ADCC, an antibody first binds to a cell-surface antigen and then recruits the immune effector cells to destroy the target cells, such as cancer cells carrying antigens recognized by antibodies. The Fc gamma receptor IIIa (FcγRIIIa) on natural killer (NK) cells binds to the Fc region of the antibody, which region has a major antibody effector function in the immune system. This binding results in lysis and apoptosis of the targeted cell by NK-cell-mediated killing. A decrease of fucosylation at Asn297 in the antibody Fc domain significantly increased antibodies’ binding affinity to FcγRIIIa and, further, improved ADCC potency [38, 39, 40].
2 Technologies for Glycoengineering Through Gene Knocking Down, In, and Out
Strategies for CHO glycoengineering include the knockdown or knockout of enzymes such as sialidase or fucosyltransferase, along with the overexpression or knocking-in of glycosylation enzymes, such as glycosyltransferases and sialyltransferases. Genetic engineering approaches, including small interfering RNA (siRNA), short hairpin RNA (shRNA), ZFN, TALEN, and CRISPR/Cas9, aim to modify gene expression [37, 41,42,43,44,45,46,47,48,49,50,51], while other methods can amplify the expression of a target gene, such as by overexpression and knockin. Both siRNA and shRNA have extensive use in decreasing gene expression, thus playing a role in both reduced fucosylation and sialidase cleavage. Table 1 compares the current technologies for ZFN, TALEN, and CRISPR/Cas9, all of which can be used to modify expression in glycosylation pathways.
As an example, fucosylation is often controlled by gene knockdown and knockout strategies. The removal of core fucose can be highly advantageous for improving the therapeutic efficacy of mAbs. The core fucosylation is defined by the transferring of fucose from GDP-fucose to GlcNAc in an α-1,6 linkage catalyzed by an α-1,6-fucosyltransferase (encoded by α-1,6-fucosyltransferase [FUT8]). In one study, overexpression of GnT-III was able to compete with native fucosyltransferase and produce a afucosylated antibody [52]. The results in that study indicated that GnT-III inhibited the core FUT8, increasing the production of a bisected afucosylated antibody with enhanced ADCC activity [52]. Coexpression of GnT-III with Golgi alpha-mannosidase II (ManII) resulted in more complex oligosaccharides compared with the expression of GnT-III alone [52]. The overall results indicate the importance of decreased fucosyltransferase activity for improving ADCC. A number of strategies can be implemented to lower or silence fucosyltransferase activity, including siRNA, shRNA, ZFN, TALEN, and CRISPR/Cas9 [37, 41,42,43,44,45,46,47,48,49,50,51].
2.1 siRNA
siRNA can be used as a transient or a stable method to suppress specific gene expression using RNA interference. Two siRNA sequences were found that reduced the expression of FUT8 in CHO DG44 cells to 20% of the level in parental controls [45]. The decrease in mRNA expression corresponded to a 40% fucosylated antibody with 100 times the ADCC of that for control cells [45]. Additionally, clone stability was demonstrated, as the ability to produce antibody with decreased fucosylation continued over repeated passages and fed-batch culture [45]. Interestingly, FUT8 knockdown was more effective in the exponential phase than in the stationary phase of culture [45]. In summary, this siRNA approach did not completely knockout FUT8 expression, but the decreased expression resulted in decreased fucosylation and enhanced ADCC.
In another study, a CHO cell line, also with FUT8 knocked down using siRNA, was created and compared with two lectin-mutated defucosylation cell lines—an endogenous GDP-fucose 4,6-dehydratase (GMD)-deficient cell line (Lec13) and an endogenous GnT-1-deficient cell line (Lec1) [41, 53]. These lectin-mutated cell lines produced afucosylated antibody, but over culture time, the percentage of fucosylated antibody increased [41]. In contrast, in this study, the FUT8 siRNA cell line produced completely afucosylated antibody throughout cell culture [41]. Subsequent scaling of the experiment to bioreactors with pH and dissolved oxygen control yielded similar results, including afucosylated antibody and enhanced ADCC, from the FUT8 siRNA cells [41]. Thus, siRNA is an important tool for controlling fucosylation at different scales in bioprocess development.
Moreover, three key enzymes in the fucosylation pathways in CHO cells have been identified, and knockdown of these key enzymes—FUT8, GDP-fucose transporter (GFT), and GDP-fucose 4,6-dehydratase (GMD)—using separate siRNA vectors, has also been achieved to study the effect on fucosylation of recombinant glycoproteins. Both the FUT8 and GMD siRNA cell lines were separately found to produce afucosylated antibodies [37]. In contrast, knockdown of 98% GFT expression at the mRNA level yielded only 40% reduction of the Fc fucosylated oligosaccharide [37]. After it was demonstrated that GMD inhibition with siRNA removed intracellular GDP-fucose and yielded afucosylated antibodies, it was shown that GMD-KO CHO DG44 cells produced fucosylated antibodies upon medium supplementation of l-fucose during culture [37]. Cell culture samples were obtained and the level of UDP-glucose and the oligosaccharide profiles were determined with high-performance liquid chromatography (HPLC) and matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (MS), respectively [37]. These findings highlight that the degree of fucosylation can be controlled through both cell engineering and media manipulation.
In related experiments, knockdown of FUT8, GFT, or GMD resulted in low levels of fucosylated antibody [43]. Furthermore, the combined knockdown of FUT8 and GMD, using siRNA, synergistically improved the fraction of afucosylated antibody [43]. From these results, a tandem expression vector was designed to simultaneously knockdown FUT8 and GMD expression in CHO cells [43]. This strategy produced completely afucosylated antibody at constant levels during passaging and adaptation to serum-free medium for 2 months [43]. This result highlights that combined knockdown of fucosylation enzymes can result in antibodies that are completely devoid of fucosylation.
2.2 shRNA
One disadvantage of siRNA is the quick degradation of the RNA in the cell. shRNA is more stable because, following transfection, the shRNA becomes an active double strand. Using an alternative strategy,CHO DG44 cells transfected with FUT8 shRNA showed less than 5% FUT8 mRNA expression, which resulted in the production of 12% fucosylated antibody and enhanced ADCC compared with that in the parental cells [42]. Glycoform profiles were determined by electrospray ionization mass spectrometry (ESI-MS) [42]. One benefit of shRNA over siRNA technology is the extended efficacy of the former. Stability studies demonstrated that FUT8 knockdown was maintained for over 4 weeks [42]. After prolonged culture, the mRNA expression of FUT8 and the percentage of fucosylated antibody remained consistently low [42]. Thus, it is possible to achieve significant reduction in antibody fucosylation using either siRNA or shRNA.
2.3 Random Mutagenesis and Homologous Recombination Knockout Selection
Originally, the knockout of genes such as FUT8 required the screening of numerous clones to find one in which the gene had been randomly mutated. Sequential homologous recombination was used to knockout both alleles of FUT8 [44]. Gene targeting by homologous recombination is a useful strategy to genetically modify any chosen allele in a predetermined way without affecting any other locus in the genome [54]. This strategy produced completely afucosylated antibodies, with the growth and viability of the cell culture being similar to that in the parental controls [44]. Assays to determine binding activity, ADCC, and complement-dependent cytotoxicity (CDC) revealed no effect on the binding activity or CDC of the FUT8−/− knockout, whereas the ADCC was increased 100-fold over that of a commercial antibody, Rituxan (rituximab; Genentech), without the FUT8 knockout [44]. The FUT8−/− knockout showed significantly stronger binding to FcγRIIIa than the parental FUT8+/+ antibodies [44]. Additionally, knockout of one or both alleles of FUT8 was compared and it was found that a hemizygous FUT8+/− knockout did not reduce fucosylation completely [44]. Thus, knockout of both FUT8 alleles can be used as a strategy to produce completely afucosylated antibody therapeutics from CHO cells.
Mutants can also be used to understand glycosylation and identify new targets for intervention. Treating CHO cells with the cytotoxic lectin Ricinus communis agglutinin I (RCA-I), which is specific for terminal beta-1,4-linked galactose [55], was designed to select mutants with defects in the N-glycosylation pathway upstream of galactose addition. Surprisingly, RCA-I-resistant CHO mutants contained mutations in the N-acetylglucosaminyltransferase I (GnT-I) gene similar to those in the Lec1 mutant [56]. Possibly, RCA-I may not be specific for terminal beta-1,4-linked galactose, and may bind other glycan structures, except for Man5GlcNAc2 [57]. Without functional GnT-I, cells fail to transfer GlcNAc to Man5GlcNAc2. By restoring functional GnT-I in these mutants, the sialic acid content of recombinant proteins in transient expression and stably transfected clones increased [56]. While the molecular mechanism for this phenomenon remains unknown [58], recombinant EPO generated in the RCA-I-restored mutant cell line with GnT-1 exhibited an increase in sialylation of 30% over the control [59]. In addition, the percentage of tri- and tetra-antennary glycans on EPO produced by the GnT I-restored CHO-GnT I-deficient cells increased, as measured by MS [59].
2.4 ZFNs
An alternative to random mutagenesis is to apply ZFN technology. Zinc fingers are transcription factors that recognize three to four bases of a sequence and can be used to target a specific sequence. ZFNs contain the zinc finger domain and FokI endonuclease domain, which must dimerize for activity that ensures specificity [60]. In one of the initial applications, ZFNs were designed to eliminate FUT8 function [46]. The benefit of this technique is the applicability of the created ZFNs to any CHO cell line [46]. The technology allows for targeting point mutations with in-frame, short deletions [46]. Zinc finger-transfected cells had growth, antibody productivity, and glycosylation patterns similar to those in the parental controls; however, the antibodies produced were completely afucosylated [46].
In another experiment, ZFNs were used to generate CHO cell lines deficient in mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase (GnT-I) [47]. This resulted in the production of glycoproteins with high Man5 species [47]. Using ZFNs reduced off-target effects and maintained the same growth and productivity as that in the parental cell line, thus demonstrating process robustness and potential for scale-up [47]. An application of this knockdown is to generate mannose receptor targeted biologics. ZFNs offer an alternative to generating knockouts and may have important applications in future cell engineering strategies to control glycosylation.
2.5 TALENs
Another novel technology for genetic engineering is TALENs. This technology is more flexible than ZFN technology, because TALENs are dimeric transcription factor nucleases—composed of 33–35 amino acid modules—that can each target a single nucleotide [60]. Many companies offer custom design of TALENs, thus reducing the cost of TALENs compared with that of ZFNs. In recent research, knockout of CHO FUT8 via the simultaneous TALEN-mediated integration of an antibody cassette was demonstrated, and this process produced afucosylated antibodies [61]. Another novel technology applied TALEN and precise integration into target chromosome (PITCh) vector-mediated integration of long gene cassettes in CHO cells [48]. Results showed over 9 kb whole plasmid integration and over 7 kb backbone-free integration at the defined genomic locus, and the production of a recombinant single-chain Fv (variable region)-Fc(constant region) protein [48]. The method demonstrated the applicability of TALENs for high-throughput knockin of large DNA into CHO cells. Thus, TALENs can serve as a beneficial tool for biotechnology applications, such as improving the generation of high-producing cell lines with desirable glycosylation.
2.6 CRISPR/Cas9
Finally, CRISPR/Cas9 represents one of the newest and most rapidly expanding methods for genome engineering in CHO cells. First, Cas9 generates a double-strand DNA break at a site determined by the guide RNA; the system is different from those of ZFNs or TALENs because it relies on bacterial adaptive immunity [60]. Multiple guide RNAs can be used to facilitate simultaneous mutations, and the system can be applied to activator or repressor domains to control gene expression [60]. One potential disadvantage, however, is the shorter length of the guide RNA compared with ZFNs and TALENs, which could result in off-target effects [60]. The first published results of CRISPR/Cas9 genome editing in CHO cells demonstrated the successful disruption of C1GALT1 specific chaperone 1 (COSMC) and FUT8 [49]. The single guide RNAs generated an indel frequency of 47.3% in COSMC and 99.7% in FUT8 (with lectin selection) [49]. In addition, the bioinformatics tool CRISPy was established to identify the single guide RNA sequences in the CHO genome [49]. In related research, CRISPR/Cas9 was used to simultaneously disrupt FUT8, BCL2 antagonist/killer, and BCL2 associated X in CHO cells [50]. Single cell sorting revealed that, among 97 clones, there were 34 triple-, 23 double-, and four single-disrupted cell lines [50]. The triple-disrupted clones were confirmed to have removal of BAK and BAX, as well as decreased fucosylation [50]. Additionally, the disrupted cell lines were more resistant to apoptosis than the parental cells [50]. Further, instead of targeting the FUT8 gene, the knockout of key enzymes in fucosylation pathways provided alternatives to suppress fucosylation, such as knockout GDP-d-mannose-4,6-dehydratase (GMD) and GDP-4-keto-6-d-deoxymannose epimerase/GDP-4-keto-6-l-galactose reductase (FX), which are involved in the de-novo synthesis of GDP-fucose [62]. Disruption of both alleles of the FX gene via CRISPR/Cas9 led to the expression of an antibody with fully afucosylated glycan profiles [62]. CRISPR/Cas9 can thus serve as a useful tool for glycoengineering, because of its potential to affect multiple genes involved in glycosylation, sialylation, and fucosylation. These successes highlight the applicability of CRISPR/Cas9 for genome editing.
Recently, the combination of ZFNs, TALENs, and CRISPR/Cas9 was used for CHO glycoengineering to inactivate the GDP-fucose transporter and improve ADCC [51]. Mass spectrometry was used to identify that the EPO-Fc and anti-Her2 antibody produced in the modified cell lines lacked core fucosylation [51]. Removal of the core fucose did not affect cell growth or productivity as compared with these properties of the parental cell lines [51]. This experiment shows that genome editing techniques are applicable to CHO glycoengineering and can provide results that aid bioprocess development.
3 CHO Glycoengineering
CHO glycoengineering efforts aim to alter glycosylation steps by either increasing or decreasing specific glycan attachment, including terminal sialylation, or, alternatively, by reducing the cleavage of sialic acid by sialidase. The previous section highlighted the efforts made to reduce core fucosylation by gene knockdown and knockout. Glycoengineering strategies are described in this section, including the overexpression of glycosyltransferases, overexpression of galactosyltransferases, overexpression of sialyltransferases, and reduction in sialidase cleavage. These modifications also include efforts to alter terminal sialylation by overexpression or, alternatively, by reducing the cleavage of sialic acid by sialidase.
3.1 Overexpression of GnT Genes
During N-glycosylation, various monosaccharides are added to the oligosaccharide chains of glycoproteins. One strategy to improve glycosylation was through the overexpression of rat GnT-III in CHO DG44 cells producing recombinant antibody [63]. Glycan analysis by HPLC revealed that most glycoproteins displayed bisecting GlcNAc residues [63]. This resulted in a 10- to 20-fold improvement in ADCC, as determined by the increased affinity of the antibody to Fcγ receptor III (FcγRIII), without affecting cell growth or antibody productivity [63]. Similarly, increased expression of GnT-III increased the bisecting GlcNAc residues [64,65,66]. Improving the proportion of glycans that have GlcNAc residues has positive effects on therapeutic efficacy. Additionally, it has been shown that GnT-III competes with beta-1,4-galactosyltrasferase; as bisecting GlcNAc residues increase, there is a concomitant decrease in the complexity of the glycans [64,65,66]. Even more importantly, GnT-III will compete with the core FUT8 enzyme, which leads to decreased fucosylation, while increasing bisecting GlcNAc residues. Ultimately, the reduction in fucosylation may be the primary reason for the increased ADCC observed in the CHO cells that overexpress GnT-III. Recently, GnT-III was coexpressed with fucosyltransferase 7 in order to optimize glycoengineering by localizing the glycosyltransferase in the Golgi machinery [67]. The approach was able to control the N-glycans with defined structural motifs; the addition of bisecting GlcNAc, as measured by HPLC and MS, resulted in an increased ADCC for the therapeutic agent cetuximab [67].
GnT-IV and GnT-V are involved in multiantennary glycan formation [64,65,66]. Overexpression of branching genes can increase complexity, as well as increasing sialylation acceptor sites. Shown in Fig. 3 are examples of bi-, tri-, and tetra-antennary structures. The structures include complex-type N-glycans with GlcNAc that can be extended to contain the disaccharide Gal-beta-1,4-GlcNAc, sometimes capped by a terminal sialic acid. The formation of tri- and tetra-antennary N-glycans is controlled by the enzymatic actions of GnT-IV and GnT-V. Cell proliferation, cell-surface signaling [23], cancer metastasis, regulation of T-cell activation [68], and the rate of therapeutics clearance by the kidneys are all affected by the actions of GnT-IV and GnT-V [69]. In one study, only a small fraction of glycoproteins produced in a CHO cell line contained GlcNAc beta-1-6 branching controlled by GnT-V [66]. This suggested genetic engineering approaches targeting GnTs might serve to improve the production of recombinant therapeutics. Overexpression of GnT-IV or GnT-V individually was found to increase the antennarity of the glycoform profile, as determined by an increase in reactivity with Datura stramonium agglutinin [70] lectin blot [64,65,66].
In order to control the multi-antennary glycoforms of recombinant proteins, the overexpression of GnT-IV and GnT-V was used in CHO cells producing human interferon (IFN)-gamma and EPO [66, 71]. In both cases, tri- and tetra-antennary sugar chains comprised more than 50% of the total sugar chains [71]. At the same time, this resulted in higher levels of poly LacNAc [66, 71]. In another study, mouse ST3 and/or rat ST6 were incorporated into CHO cell lines stably transfected with GnT-V that were producing IFN-gamma [65]. Results showed that over 60% of the glycoforms were sialylated with alpha-2,3- and alpha-2,6-linkages [65].
Recently, a combined approach was used to increase both branching and sialylation in CHO-K1 cells producing EPO [71]. Both GnT-IV and GnT-V, as well as human alpha-2,6-sialyltransferase (ST6Gal1) were incorporated in the CHO-K1 cells, resulting in a pool of 92% N-glycans with tri- and tetra-antennarity [71]. This also improved sialylation, as measured by an increase of 45% in tetra-sialylation [71]. The approach showed that combining the genetic integration of complementary genes could significantly enhance glycosylation branching complexity, as well as enhancing overall improvements in sialylation.
O-linked glycosylation can also be modified through cell engineering approaches. Although studies of O-glycosylation are limited, there are important biological applications of O-glycans. During O-glycosylation, various carbohydrate chains are added to the serine or threonine residues of proteins. Cell engineering strategies have attempted to control O-glycosylation by altering GnT activity. In one experiment, the core 2 beta1-6GlcNAc transferase (C2GnT) was overexpressed in CHO DG44 cells [72]. The increase in enzyme activity was hypothesized to play a role in T-cell activation and immunodeficiency [72]. In another study, the combined overexpression of C2GnT and the knockdown of CMP-sialic acid: Gal-beta-1,3-GalNAc-alpha-2,3-sialyltransferase (ST3Gal1) was evaluated in CHO-K1 cells [73]. ST3Gal1 inhibition was predicted to redirect O-glycosylation toward the production of tetrasaccharide structures important for cell-cell interaction [73]. This experiment suggests that cell engineering can be used to simultaneously upregulate and downregulate competing enzymes involved in glycosylation.
Recently, extended C1 beta-3 GnT-III, C2 beta-3 GnT-I, and C3 beta-3 beta-1,4-N-acetylglucosaminyltransferase VI were transiently transfected into CHO cells and the resulting O-glycome was mapped by MS [39]. This transfection experiment resulted in extended core 1 and core 3 O-glycans, as well as the increased expression of core 2 O-glycans [39]. Overall, these results suggest that cell engineering can be applied to O-glycosylation in order to control the branching of glycans. This will aid bioprocess developments to generate mucin-type recombinant proteins.
3.2 Overexpression of Sialyltransferase and Galactosyltransferase Genes
Glycoengineering by increasing the expression of sialyltransferase enzymes has been an effective strategy to control sialylation; these enzymes add the sialic acid (Neu5Ac) residue to the terminal galactose. There are six beta-galactoside alpha 2,3-sialyltransferases (ST3GAL1-6) and two beta-galactoside alpha-2,6-sialyltransferases (ST6GAL1-2) that generate terminal sialic acids in mammalian cells. Whereas human glycoproteins contain both alpha-2,3- and alpha-2,6-linked sialic acid, CHO cells natively contain almost exclusively alpha-2,3-linked sialic acid on their glycoproteins. This means that efforts to generate more human-like glycoforms can be implemented in CHO cells. As stated above, normally, CHO cells produce almost exclusively alpha-2,3-linked sialic acid, whereas in humans, glycoproteins represent a pool of alpha-2,3- and alpha-2,6-linked sialic acid. Rat alpha-2,6-sialyltransferase was transfected into CHO cells producing tissue plasminogen activator (tPA) and it was observed that competing glycosyltransferases yielded glycoproteins with different sialic acid linkages [74]. Thus, recombinant proteins with a mixture of alpha-2,3- and alpha-2,6- sialic acid can be generated; this mixture is similar to the pool of sialylated proteins in humans.
A combination of ST3GAL3, ST3GAL4, and ST3GAL6 knockdown using siRNA has revealed that all three enzymes are involved in alpha-2,3-sialylation in CHO cells [33]. Of these enzymes, ST3GAL4 was the most critical for glycoprotein alpha-2,3-sialylation [33]. In contrast, in humans, ST6GAL1 prefers the Gal beta-1-4GlcNAc disaccharide sequence linked to a protein, whereas ST6GAL2 prefers free disaccharide Gal beta-1-4GlcNAc substrate [75].
Lee et al. found that competition between endogenous alpha-2,3-sialyltransferase and heterologous alpha-2,6-sialyltransferase yielded glycoproteins with alpha-2,3- and alpha-2,6- linkages in CHO cells [76]. As the expression of alpha-2,6-sialyltransferase increased, enzymatic assays revealed only a slight increase in total sialyltransferase activity in transfected cells; of this activity, 50% was correlated to alpha-2,6-sialyltrasferase [76]. Furthermore, the transfected cells attached alpha-2,6-sialic acid to 20% of terminal galactose [76]. Another group found that the expression of human alpha-2,6-sialyltransferase in CHO cells [77] resulted in an increased percentage of tri- (by 8%) and tetra- (by 16%) sialylated recombinant thyroid-stimulating hormone [77]. The increase in more fully sialylated protein did not affect hydrophobicity or bioactivity [77]. This research indicates the potential for more human-like sialylation of recombinant therapeutics.
Another means to increase sialylation is to make more sites available for adding sialic acid. This can be achieved by overexpressing human β 1,4-galactosyltransferase in CHO cells in order to reduce the oligosaccharides terminating with GlcNAc [68]. In one study, the overexpression of human β 1,4-galactosyltransferase in CHO cells significantly reduced oligosaccharides terminating with GlcNAc compared with results in controls [78].
The engineering of galactosyltransferases is frequently used in combination with sialyltransferase engineering. In order to increase the level of sialylation, the enzyme alpha-2,3-sialyltransferase was overexpressed along with beta-1,4-galactosyltransferase in CHO cells [78]. Similarly, the coexpression of alpha-2,6-sialyltransferase with beta-1,4-galactosyltransferase effectively increased sialic acid content [79]. Results indicated that the overexpression of galactosyltransferase improved the homogeneity of glycoforms, while the overexpression of sialyltransferase improved the sialylation of recombinant protein to 90% compared with the level in parental cells [78]. The effect of increased sialylation was verified in rat models, where it was shown that recombinant proteins with increased sialic acid had increased circulation time [78]. Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) MS was used to detect charged and neutral oligosaccharides in negative and positive ion modes, respectively [78]. Despite overexpression, glycoproteins that were not fully sialylated were detected, a finding which may be attributed to sialidase cleavage or steric hindrance [78]. In another study, Jeong et al. studied the effect of the overexpression of beta-1,4-galactosyltransferase and alpha-2,3-sialyltransferase in CHO cells producing recombinant EPO [80]. The coexpression of galactosyltransferase and sialyltransferase resulted in an increase in sialic acid content, from 6.7 to 8.2 mol of sialic acid per mole of EPO, and an increase in trisialylated glycans from 17.3% to 35.5% compared with levels in parental cells [80]. At the same time, cell growth, metabolism, and protein productivity were not affected [80]. There was virtually no change in tetrasialylated glycans, suggesting possible steric hindrance in attaching a fourth CMP-sialic acid to the trisialylated glycans or sialyltransferases having branch specificity [80]. This result highlights the importance of both galactosyltransferase and sialyltransferase in producing homogenous, sialylated glycoproteins. Both enzymes are important for maintaining the lot-to-lot consistency of glycoprotein therapeutics, which is required for consistent manufacturing and drug efficacy.
The studies cited above [78, 79, 80] showed the effect of sialyltransferase and galactosetransferase expression on the sialylation of glycoproteins. However, transporting CMP-sialic acid to the Golgi apparatus is a potential bottleneck, owing to the levels or activity of the CMP-sialic acid transporter (CMP-SAT) that transports CMP-Neu5Ac into the Golgi apparatus. Overexpression of CMP-SAT alone resulted in a 4–16% increase in the site sialylation of IFN-gamma [81]. Following these findings, combinatorial efforts have sought to engineer multiple genes in the pathway in order to improve sialic acid content in the intracellular pool and improve the transport of sialic acid substrates in the Golgi apparatus.
Another approach is to implement methods that increase the levels of the sialylation substrate, CMP-Neu5Ac (or CMP-sialic acid). In order to enhance both activities, human alpha-2,3-sialyltransferase and CMP-sialic acid synthase were simultaneously overexpressed in CHO cells producing recombinant EPO [82]. This resulted in increased sialylation; however, the increase was attributed to alpha-2,3-sialyltransferase alone [82]. Coexpression was found to increase the pool of intracellular CMP-sialic acid, suggesting that a bottleneck to sialylation is the transport of sialic acid into the Golgi apparatus [82]. Following this finding, alpha-2,3-sialyltransferase, CMP-sialic acid synthase, and CMP-sialic acid transporter were simultaneously overexpressed and there was an additional increase in tri- and tetra- sialylated glycans concomitant with a decrease in monosialylated glycans [82]. This result highlights how genetic engineering strategies can be used to overcome pathway bottlenecks in both the generation of glycosylation substrates and the transfer of the substrate to the oligosaccharide target, by using a transferase enzyme in order to maximize the sialylation of recombinant proteins.
In another experiment, the enzymes uridine diphosphate-N-acetyl glucosamine 2-epimerase/N-acetyl mannosamine kinase (GNE/MNK), CMP-sialic acid transporter, and alpha-2,3-sialyltransferase were simultaneously introduced in CHO cells producing recombinant EPO [83]. GNE/MNK initiates sialic acid biosynthesis; a mutant variant of the enzyme was used in order to eliminate feedback control by the end product of the pathway. Subsequently, CMP-sialic acid transporter sequesters CMP-sialic acid into the Golgi apparatus, where sialyltransferase then adds sialic acid to the maturing glycoprotein. Results indicated that the sialic acid content of recombinant EPO increased by 43% compared with that in parental cells; additionally, there was a 32% increase in tetrasialylated EPO and declines of 50% in both monosialylated and asialylated EPO [83]. This study provides further evidence that the combined simultaneous transfection of multiple enzymes in the sialic acid biosynthetic and transfer pathways can have a significant impact on overall product sialylation.
3.3 Inhibition of Sialidase Activity
Sialidases are exoglycosidases that catalyze the hydrolytic removal of sialic acid from sialoglycoconjugates (glycoproteins, polysaccharides, gangliosides) [84]. Four sialidases (Neu 1–4) have been identified in human, mouse, rat, and CHO cells, and their activity is localized to different subcellular compartments: Neu1 is located in the lysosome, Neu2 is located in the cytoplasm, Neu3 is located in the plasma membrane, and Neu4 is also located in the lysosome [84,85,86]. Thus, sialidase function varies as a result of the different substrate specificities and subcellular locations [32]. Sialidase cleavage occurs in cell culture as viability decreases, and this cleavage leads to the desialylation of recombinant glycoproteins [32, 85].
In order to decrease sialidase activity, a CHO cell line was developed that expressed sialidase antisense RNA [87]. Sialidase activity in this cell line was reduced by 40%, compared with the control culture; this reduction corresponded to an increase in sialic acid content ranging from 20 to 37% [87]. Over the culture duration, sialidase concentration increased in both the control and antisense cultures [87]. However, the sialidase level in the antisense culture remained 40% lower than that in the control cells [87]. Another important finding was the consistent viability between the control and antisense cultures, which suggests that sialidase antisense RNA is a useful strategy for reducing sialidase cleavage [87]. The finding that sialidase antisense RNA expression was not completely knocked out indicated the likelihood that no severe effects on growth or metabolism had occurred. This result highlights the capacity to manipulate sialidase levels as a means to maintain sialylated glycoforms for recombinant protein production.
In another experiment, Neu2 knockdown was used to decrease cytosolic sialidase activity in CHO cells producing IFN-gamma [85]. After siRNA sequences were compared, the most active sequence was transfected into CHO cells, resulting in a reduction in sialidase activity of 60% relative to control cells [85]. Cell culture glycan samples were analyzed by MS and it was found that reducing sialidase cleavage did not affect the glycan site distribution [85]. This finding is important for the development of the cell culture process so that consistent batches of recombinant protein are produced. For one clone with decreased sialidase cleavage, there was no change in the percentages of asialoglycans, monosialylated glycans, or disialylated glycans [85]. However, over the duration of the control cell culture, there was a decrease in disialylated glycans, with a concomitant increase in asialoglycans and monosialylated glycans [85]. RNAi knockdown of Neu2 did increase sialic acid content, but only when cells were in the death phase [85, 87, 88]. Although sialic acid content does not always increase during the growth phases with sialidase RNAi, it is possible to maintain consistent glycoforms and prevent the desialylation of glycoforms in later culture stages with this strategy.
In another study, siRNA and shRNA were used to knockdown Neu1 and Neu3 sialidase genes [89]. Reduced expression of Neu3 resulted in a 98% reduction in Neu3 sialidase activity in CHO cells, corresponding to increases in sialic acid content of 33% and 26% for samples from the cell stationary phase and death phase, respectively [89]. Interestingly, application of the siRNA technique to knockdown Neu3 (located in the plasma membrane) individually resulted in negligible sialidase activity, whereas knockdown of Neu2 (located in the cytoplasm) individually only reduced sialidase activity to 40% of the control level [32]. Unlike Neu2 knockdown effects that acted exclusively in the death phase, protein sialylation was increased throughout cell culture by Neu3 knockdown, suggesting different mechanisms of sialylation control by Neu2 and Neu3, respectively [32].
In summary, many strategies, involving both the upregulation and downregulation of enzymes involved in the glycosylation pathways, can be manipulated to control cellular glycosylation. Some of the approaches described in this chapter are highlighted in Fig. 4, which shows the effect of specific gene overexpression or the knockdown of enzymes involved in various glycan processing steps, including sialylation and fucosylation. In the next section, we introduce the importance of a systems biology approach to understand glycosylation and to elucidate glycan compositions that can be used to drive genetic engineering strategies in the future.
Examples of different glycoengineering targets. Strategies for glycoengineering include efforts to increase branching, increase sialylation and galactosylation, and decrease fucosylation. Enzyme targets are shown for different strategies and are outlined in red if expression is typically increased or green if expression is typically decreased. UDP uridine diphosphate,… Cytidine monophosphate (CMP)
4 CHO Glycoproteomics and Combined ‘Omics’
Since glycan patterns are exposed on cell surfaces, they are ready targets for high-throughput technologies such as glycoproteomics [90, 91]. Indeed, the development of sophisticated analytical techniques [92,93,94,95] and data analysis tools [96,97,98,99,100] provides increasing opportunities to utilize high-throughput screening for glycans as disease markers and for the structural classification of therapeutic proteins. Glycogene microarrays, lectin chips, and RNA sequencing tools are widely used to analyze the whole glycogenome and the changes in glycosylation enzymes, as shown in Fig. 5. In addition to these tools, recent advances in MS)enable qualitative and quantitative analyses of glycans, glycosites, glycopeptides, and intact glycoproteins [101].
Methods for glycan analysis. New advances in sample preparation and analysis enable the identification and quantification of glycoproteins with high accuracy and reproducibility. Examples include lectin microarrays, ultra-performance liquid chromatography (UPLC), and liquid chromatography tandem mass spectrometry (LC/MS/MS). Abbreviations: Asn asparagine, Ser serine, Thr threonine, HCD higher-energy collisional dissociation
4.1 Glycoproteomics
Glycoproteomics, a field that evaluates glycosylated proteins and their glycosylation sites [102], involves glycoprotein enrichment of the samples followed by sophisticated proteomics methods, advanced MS techniques, and powerful bioinformatics tools. Label-free quantification [103], stable isotope labeling (SILAC) [104], isobaric tag for relative and absolute quantitation (iTRAQ) [105], and tandem mass tags (TMT) [106] are some of the methods that can be used to interpret the differential expression of glycoproteins between samples, such as different clones or changing process conditions.
Solid phase extraction of glycosylated peptides (SPEG) enables the identification of N-linked glycoproteins using hydrazide chemistry. In this method, a protein mixture is equilibrated with a hydrazide resin, which binds to the carbohydrate moieties on the glycoproteins. Then, polypeptides are oxidized and enzymatically removed by peptide-N-glycosidase F for liquid chromatography tandem mass spectrometry (LC-MS) analysis [107]. A previous CHO proteome analysis used a label-free approach to identify 6,164 total proteins and glycoproteins [108]. Of these, the SPEG method revealed that at least 1,292 proteins were N-glycosylated [108]. In recent years, more developments have been made to improve the identification and quantification of glycoproteins. Glycan quantification using isobaric tags, such as aminoxyTMT and iART, is difficult owing to their tertiary amine structure [109]. A novel MS-based technology, called quaternary amine-containing isobaric tag for glycan (QUANTITY), was recently developed to improve the complete labeling of glycans and increase reporter ion intensity upon second stage of mass (MS2) fragmentation [109]. The QUANTITY labeling approach has been coupled with solid-phase immobilization techniques for the glycomic comparison of CHO cells engineered with glycosyltransferases [109]. Samples are first denatured and immobilized on AminoLink resin (Thermo Fisher Scientific). To stabilize sialic acid groups, p-toluidine can be used with a carbodiimide coupling reagent, and then PNGaseF releases N-glycans from the solid support. Next, the aldehyde group of the GlcNAc at the reducing end of the glycans from each sample can be labeled with QUANTITY, followed by an analysis with liquid chromatography tandem mass spectrometry (LC/MS/MS). A global proteomics analysis can also be conducted by performing on-bead digestion [109].
Site-specific glycan occupancy and alterations in glycoproteins are also significantly important for bioprocess development. Previously, glycosites, glycopeptides, and glycans were studied separately owing to difficulties with simultaneous analysis. Solid-phase extraction of N-linked glycans and glycosite-containing peptides (NGAG) can simultaneously analyze glycans, glycosites, and glycopeptides from complex samples [110]. First, peptides are immobilized using an aldehyde-functionalized solid support. Then, PNGaseF and endoproteinase Asp-N digestions release the N-glycans and N-glycopeptides, respectively, through enzymatic cleavage. After MS analysis, a sample-specific intact glycopeptide database is created to document the glycosites and glycans [110]. At the same time, intact glycopeptides are isolated and run by MS. The spectra are subsequently mapped to a glycosylation-specific database using GPQuest software [110, 111].
Finally, methods have recently been developed to improve our understanding of O-glycosylation. A microwave-assisted beta-elimination method has been optimized to analyze O-glycans from cells, tissues, serum, and formalin-fixed paraffin-embedded tissues [112]. In summary, the use of ‘omics’ has expanded our ability to elucidate glycan structures, glycosites, and glycopeptide composition in order to understand the glycoproteome and glycoform profiles from CHO cell cultures. These efforts seek to identify deficiencies in glycosylation profiles that may be overcome through genetic engineering intervention.
4.2 Combined ‘Omics’
More recent efforts have combined glycoproteomics with other ‘omics’ technologies for the validation and improved understanding of glycosylation. In one approach, genome-wide association studies were combined with high-throughput HPLC analysis of plasma proteins from 2,705 individuals, to reveal polymorphisms in FUT6 and FUT8, as well as those in hepatocyte nuclear factor 1-alpha (HNF1-alpha) [113]. The analysis was extended to 3,533 individuals to identify polymorphisms in MGAT5 and B3GAT1 and the protein pump SLC9A9 [114]. Another study combined epigenomics with proteomics to show that global changes in the DNA methylation of ovarian cancer epithelial cells could affect glycans by reducing core fucosylation, increasing branching, and increasing sialylation [115]. Altered expression of fucose biosynthetic genes and increased expression of MGAT5 were found to modify the branching and sialylation of secreted glycans [116]. These studies demonstrate how epigenomics and glycan structural analysis can be combined to study the effects of genes and pathways in human glycosylation that may also be important for CHO glycosylation processing.
In another approach, pathway mapping was used to correlate transcriptional regulation and glycan expression [117]. Increased polysialylation and alpha-Gal termination were observed in differentiated cell types, whereas alpha-Gal capped glycans were more abundant in extra-embryonic endodermal cells [117]. Another integration study mapped microRNA (miRNA) regulators onto glycan biosynthetic pathways by the introduction of glycomics data. Lectin microarrays were used to mimic miRNAs, enabling miRNA regulators of high mannose, fucose, and beta-GalNAc networks to be determined [6].
Finally, N-glycan and glycogene expression during the epithelial-to-mesenchymal transition was studied using a systems glycobiology approach [118]. Fucosylation and bisecting GlcNAc glycans were significantly decreased during the transition, whereas levels of high mannose type N-glycans were increased [118]. In this way, the integration of ‘omics’ tools has led to the improved understanding of how glycogene expression is controlled at genomic, transcriptomic, proteomic, and epigenomic levels.
5 Conclusions and Outlook
This review has highlighted the role of glycosylation as a critical quality attribute in the production of biotherapeutics, and more importantly it has highlighted how these glycans can be manipulated in CHO expression systems through cell engineering, as summarized in Table 2. Mammalian cell lines such as CHO can produce valuable recombinant proteins that can be accepted by humans as therapeutics. However, subtle differences exist between glycosylation in humans and other mammals, and understanding these differences requires knowledge of the physiological characteristics of each cell type. Efforts to exert control over protein glycosylation in CHO cells have been made by maximizing terminal sialylation through the overexpression of N-acetylglucosaminyltransferases, the overexpression of galactosyltransferases, the overexpression of sialyltransferases, the inhibition of sialidases, and the manipulation of CMP-sialic acid pathways. Equally important have been approaches to limit fucosylation through the overexpression of inhibiting N-acetylglucosaminyltransferases such as GnTIII, suppressing fucosyltransferase activity, and blocking the generation of the GDP-fucose substrate. The increasing use of advanced technologies such as ZFN, TALEN, and more recently CRISPR/Cas9, will greatly facilitate efforts to insert precise modifications of the glycosylation pathways into the CHO genome in future. Indeed, recent efforts have achieved comprehensive knockdown of multiple glycosyltransferases in order to control N-linked glycosylation in CHO cells [119]. This approach allows users to tailor the design of glycosylation for specific glycan profiles on recombinant glycoproteins. Furthermore, combinatorial glycoengineering approaches, including knockdowns, knockouts, knockins, and knockups, will be increasingly implemented to overcome multiple interacting pathway bottlenecks. These tools will enable highly refined and targeted modifications to be made to the processing capability of CHO cells in order to meet the need for flexible production capabilities, as well as meeting the need for the highly specified glycan targets required in biosimilar generation. Finally, the generation of ‘omics’ data sets is propelling a systems biology revolution to increase our understanding of CHO physiology and our capacity to modify glycans in different ways. Our ability to elucidate, characterize, quantify, and finally modify glycoproteins emerging from CHO, as well as the enzyme activities present in CHO, will facilitate the development of a superior CHO production platform that will yield consistent and desirable glycoforms in the future. In the coming decades, the emerging systems glycobiology integration of glycogenomics, glycoproteomics, glycomics, epiglycogenomics, and glycoinformatics, together with our ever-expanding toolkit for genome engineering, promises to accelerate our understanding of glycosylation in CHO and other mammalian cell lines, as well as increasing our capacity to control glycan processing more effectively.
Abbreviations
- ADCC:
-
Antibody-dependent cellular cytotoxicity
- Asn:
-
Asparagine
- BHK:
-
Baby hamster kidney
- CDC:
-
Complement-dependent cytotoxicity
- CHO:
-
Chinese hamster ovary
- CMP-SAT:
-
cytidine 5′-monophosphate (CMP)-sialic acid transporter
- CRISPR:
-
Clustered regularly interspaced short palindromic repeats
- Dol-P:
-
Dolichol phosphate
- EPO:
-
Erythropoietin
- ER:
-
Endoplasmic reticulum
- ESI-MS:
-
Electrospray ionization mass spectrometry
- Fc:
-
Fragment crystallizable
- FcγRIIIa:
-
Fc gamma receptor IIIa
- FUT8:
-
α-1,6-fucosyltransferase
- FX:
-
GDP-4-keto-6-d-deoxymannose epimerase/GDP-4-keto-6-l-galactose reductase
- GFPP:
-
GDP-fucose pyrophosphorylase
- GFT:
-
GDP-fucose transporter
- GlcNAc:
-
N-acetylglucosamine
- GMD:
-
GDP-fucose 4,6-dehydratase
- GNE/MNK:
-
Uridine diphosphate-N-acetyl glucosamine 2-epimerase/N-acetyl mannosamine kinase
- GnT-1 or Mgat1:
-
N-acetylglucosaminyltransferase I
- GnT-II or Mgat2:
-
Beta-1,2-N-acetylglucosaminyltransferase II
- GnT-III or Mgat3:
-
Beta-1,4-N-acetylglucosaminyltransferase III
- GnT-IV or Mgat 4:
-
Beta-1,2-N-acetylglucosaminyltransferase IV
- GnT-V or Mgat 5:
-
Beta-1,2-N-acetylglucosaminyltransferase V
- HEK:
-
Human embryonic kidney
- HNF1-alpha:
-
Hepatocyte nuclear factor 1-alpha
- HPLC:
-
High-performance liquid chromatography
- LacNAc:
-
Acetyl lactosamine
- mAb:
-
Monoclonal antibody
- MALDI-TOF:
-
Matrix-assisted laser desorption/ionization time-of-flight
- ManII:
-
Alpha-mannosidase II
- Neu5Gc:
-
N-glycolylneuraminic acid
- NK:
-
Natural killer
- OST:
-
Oligosaccharyltransferase
- RCA-I:
-
Ricinus communis agglutinin I
- Ser:
-
Serine
- shRNA:
-
Short hairpin RNA
- siRNA:
-
Small interfering RNA
- SPEG:
-
Solid phase extraction of glycosylated peptides
- TALEN:
-
Transcription activator-like effector nuclease
- Thr:
-
Threonine
- tPA:
-
Tissue plasminogen activator
- ZFN:
-
Zinc finger nuclease
References
Ghaderi D et al (2012) Production platforms for biotherapeutic glycoproteins. Occurrence, impact, and challenges of non-human sialylation. Biotechnol Genet Eng Rev 28:147–175
Hossler P, Khattak SF, Li ZJ (2009) Optimal and consistent protein glycosylation in mammalian cell culture. Glycobiology 19(9):936–949
Lepenies B, Seeberger PH (2014) Simply better glycoproteins. Nat Biotechnol 32(5):443–445
Aggarwal RS (2014) What’s fueling the biotech engine-2012 to 2013. Nat Biotechnol 32(1):32–39
Jiménez D et al (2005) Contribution of N-linked glycans to the conformation and function of intercellular adhesion molecules (ICAMs). J Biol Chem 280(7):5854–5861
Agrawal P et al (2014) Mapping posttranscriptional regulation of the human glycome uncovers microRNA defining the glycocode. Proc Natl Acad Sci 111(11):4338–4343
Palomares LA, Estrada-Mondaca S, Ramirez OT (2004) Production of recombinant proteins: challenges and solutions. Methods Mol Biol 267:15–52
Walsh G, Jefferis R (2006) Post-translational modifications in the context of therapeutic proteins. Nat Biotechnol 24(10):1241–1252
Gavel Y, Vonheijne G (1990) Sequence differences between glycosylated and nonglycosylated Asn-X-Thr Ser acceptor sites – implications for protein engineering. Protein Eng 3(5):433–442
Stavenhagen K et al (2013) Quantitative mapping of glycoprotein micro-heterogeneity and macro-heterogeneity: an evaluation of mass spectrometry signal strengths using synthetic peptides and glycopeptides. J Mass Spectrom 48(6):i
Wang Q et al (2017) Glycoengineering of CHO cells to improve product quality. Methods Mol Biol 1603:25–44
Varki A, Schauer R (2009) Sialic acids. In: Varki A et al (eds) Essentials of glycobiology. Cold Spring Harbor, New York
Aebi M (2013) N-linked protein glycosylation in the ER. Biochim Biophys Acta 1833(11):2430–2437
Aebi M et al (2010) N-glycan structures: recognition and processing in the ER. Trends Biochem Sci 35(2):74–82
Butler M, Meneses-Acosta A (2012) Recent advances in technology supporting biopharmaceutical production from mammalian cells. Appl Microbiol Biotechnol 96(4):885–894
Swiech K, Picanco-Castro V, Covas DT (2012) Human cells: new platform for recombinant therapeutic protein production. Protein Expr Purif 84(1):147–153
Padler-Karavani V, Varki A (2011) Potential impact of the non-human sialic acid N-glycolylneuraminic acid on transplant rejection risk. Xenotransplantation 18(1):1–5
Bosques CJ et al (2015) Chinese hamster ovary cells can produce galactose-alpha-1, 3-galactose antigens on proteins (vol 28, pg 1153, 2010). Nat Biotechnol 16(10):23849–23866
Muchmore EA et al (1989) Biosynthesis of N-glycolyneuraminic acid. The primary site of hydroxylation of N-acetylneuraminic acid is the cytosolic sugar nucleotide pool. J Biol Chem 264(34):20216–20223
Chung CH et al (2008) Cetuximab-induced anaphylaxis and IgE specific for galactose-alpha-1,3-galactose. N Engl J Med 358(11):1109–1117
Butler M, Spearman M (2014) The choice of mammalian cell host and possibilities for glycosylation engineering. Curr Opin Biotechnol 30:107–112
Croset A et al (2012) Differences in the glycosylation of recombinant proteins expressed in HEK and CHO cells. J Biotechnol 161(3):336–348
Zhao Y et al (2008) Branched N-glycans regulate the biological functions of integrins and cadherins. FEBS J 275(9):1939–1948
Raju TS, Jordan RE (2012) Galactosylation variations in marketed therapeutic antibodies. MAbs 4(3):385–391
Sareneva T et al (1995) N-glycosylation of human interferon-gamma – glycans at Asn-25 are critical for protease resistance. Biochem J 308:9–14
Wright A, Morrison SL (1997) Effect of glycosylation on antibody function: implications for genetic engineering. Trends Biotechnol 15(1):26–32
Spearman M, Butler M (2015) Glycosylation in cell culture. Anim Cell Cult 9:237–258
Sola RJ, Griebenow K (2010) Glycosylation of therapeutic proteins: an effective strategy to optimize efficacy. BioDrugs 24(1):9–21
Wright A et al (1991) Antibody variable region glycosylation: position effects on antigen binding and carbohydrate structure. EMBO J 10(10):2717–2723
Angata T, Varki A (2002) Chemical diversity in the sialic acids and related alpha-keto acids: an evolutionary perspective. Chem Rev 102(2):439–469
Harduin-Lepers A et al (2001) The human sialyltransferase family. Biochimie 83(8):727–737
Wang Q et al (2015) Strategies for engineering protein N-glycosylation pathways in mammalian cells. Methods Mol Biol 1321:287–305
Chung CY et al (2015) Assessment of the coordinated role of ST3GAL3, ST3GAL4 and ST3GAL6 on the alpha 2,3 sialylation linkage of mammalian glycoproteins. Biochem Biophys Res Commun 463(3):211–215
Stencel-Baerenwald JE et al (2014) The sweet spot: defining virus–sialic acid interactions. Nat Rev Microbiol 12:739
Ashwell G, Harford J (1982) Carbohydrate-specific receptors of the liver. Annu Rev Biochem 51:531–554
Cole ES et al (1993) In vivo clearance of tissue plasminogen-activator – the complex role of sites of glycosylation and level of sialylation. Fibrinolysis 7(1):15–22
Kanda Y et al (2007) Establishment of a GDP-mannose 4,6-dehydratase (GMD) knockout host cell line: a new strategy for generating completely non-fucosylated recombinant therapeutics. J Biotechnol 130(3):300–310
Shields RL et al (2002) Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human FcgammaRIII and antibody-dependent cellular toxicity. J Biol Chem 277:26733–26740
Liu J et al (2015) O-glycan repertoires on a mucin-type reporter protein expressed in CHO cell pools transiently transfected with O-glycan core enzyme cDNAs. J Biotechnol 199:77–89
Chung S et al (2012) Quantitative evaluation of fucose reducing effects in a humanized antibody on Fcgamma receptor binding and antibody-dependent cell-mediated cytotoxicity activities. MAbs 4(3):326–340
Kanda Y et al (2007) Comparison of biological activity among nonfucosylated therapeutic IgG1 antibodies with three different N-linked Fc oligosaccharides: the high-mannose, hybrid, and complex types. Glycobiology 17(1):104–118
Beuger V et al (2009) Short-hairpin-RNA-mediated silencing of fucosyltransferase 8 in Chinese-hamster ovary cells for the production of antibodies with enhanced antibody immune effector function. Biotechnol Appl Biochem 53(Pt 1):31–37
Imai-Nishiya H et al (2007) Double knockdown of α1,6-fucosyltransferase (FUT8) and GDP-mannose 4,6-dehydratase (GMD) in antibody-producing cells: a new strategy for generating fully non-fucosylated therapeutic antibodies with enhanced ADCC. BMC Biotechnol 7(1):84
Yamane-Ohnuki N et al (2004) Establishment of FUT8 knockout Chinese hamster ovary cells: an ideal host cell line for producing completely defucosylated antibodies with enhanced antibody-dependent cellular cytotoxicity. Biotechnol Bioeng 87(5):614–622
Mori K et al (2004) Engineering Chinese hamster ovary cells to maximize effector function of produced antibodies using FUT8 siRNA. Biotechnol Bioeng 88(7):901–908
Malphettes L et al (2010) Highly efficient deletion of FUT8 in CHO cell lines using zinc-finger nucleases yields cells that produce completely nonfucosylated antibodies. Biotechnol Bioeng 106(5):774–783
Sealover NR et al (2013) Engineering Chinese hamster ovary (CHO) cells for producing recombinant proteins with simple glycoforms by zinc-finger nuclease (ZFN)—mediated gene knockout of mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase (Mgat1). J Biotechnol 167(1):24–32
Sakuma T et al (2015) Homologous recombination-independent large gene cassette knock-in in CHO cells using TALEN and MMEJ-directed donor plasmids. Int J Mol Sci 16(10):23849
Ronda C et al (2014) Accelerating genome editing in CHO cells using CRISPR Cas9 and CRISPy, a web-based target finding tool. Biotechnol Bioeng 111(8):1604–1616
Grav LM et al (2015) One-step generation of triple knockout CHO cell lines using CRISPR/Cas9 and fluorescent enrichment. Biotechnol J 10(9):1446–1456
Chan KF et al (2016) Inactivation of GDP-fucose transporter gene (Slc35c1) in CHO cells by ZFNs, TALENs and CRISPR-Cas9 for production of fucose-free antibodies. Biotechnol J 11(3):399–414
Ferrara C et al (2006) Modulation of therapeutic antibody effector functions by glycosylation engineering: influence of Golgi enzyme localization domain and co-expression of heterologous beta1, 4-N-acetylglucosaminyltransferase III and Golgi alpha-mannosidase II. Biotechnol Bioeng 93(5):851–861
Patnaik SK, Stanley P (2006) Lectin-resistant CHO glycosylation mutants. Methods Enzymol 416:159–182
Tong C et al (2011) Generating gene knockout rats by homologous recombination in embryonic stem cells. Nat Protoc 6(6):827–844
Chan KF, Goh JSY, Song Z (2014) Improving sialylation of recombinant biologics for enhanced therapeutic efficacy. Pharm Bioprocess 2(5):363–366
Goh JS et al (2010) RCA-I-resistant CHO mutant cells have dysfunctional GnT I and expression of normal GnT I in these mutants enhances sialylation of recombinant erythropoietin. Metab Eng 12(4):360–368
Iskratsch T et al (2009) Specificity analysis of lectins and antibodies using remodeled glycoproteins. Anal Biochem 386(2):133–146
Goh JS et al (2014) Producing recombinant therapeutic glycoproteins with enhanced sialylation using CHO-gmt4 glycosylation mutant cells. Bioengineered 5(4):269–273
Goh JS et al (2014) Highly sialylated recombinant human erythropoietin production in large-scale perfusion bioreactor utilizing CHO-gmt4 (JW152) with restored GnT I function. Biotechnol J 9(1):100–109
Gaj T, Gersbach CA, Barbas CF (2013) ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 31(7):397–405
Cristea S et al (2013) In vivo cleavage of transgene donors promotes nuclease-mediated targeted integration. Biotechnol Bioeng 110(3):871–880
Louie S et al (2016) FX knockout CHO hosts can express desired ratios of fucosylated or afucosylated antibodies with high titers and comparable product quality. Biotechnol Bioeng 114(3):632–644
Davies J et al (2001) Expression of GnTIII in a recombinant anti-CD20 CHO production cell line: expression of antibodies with altered glycoforms leads to an increase in ADCC through higher affinity for FC gamma RIII. Biotechnol Bioeng 74(4):288–294
Fukuta K et al (2000) Control of bisecting GlcNAc addition to N-linked sugar chains. J Biol Chem 275(31):23456–23461
Fukuta K et al (2000) Genetic engineering of CHO cells producing human interferon-gamma by transfection of sialyltransferases. Glycoconj J 17(12):895–904
Fukuta K et al (2000) Remodeling of sugar chain structures of human interferon-gamma. Glycobiology 10(4):421–430
Reinl T et al (2013) Golgi engineering of CHO cells by targeted integration of glycosyltransferases leads to the expression of novel Asn-linked oligosaccharide structures at secretory glycoproteins. BMC Proc 7(6):P84
Demetriou M et al (2001) Negative regulation of T-cell activation and autoimmunity by Mgat5 N-glycosylation. Nature 409(6821):733–739
Misaizu T et al (1995) Role of antennary structure of N-linked sugar chains in renal handling of recombinant human erythropoietin. Blood 86(11):4097–4104
Park SS et al (2012) Effective correction of experimental errors in quantitative proteomics using stable isotope labeling by amino acids in cell culture (SILAC). J Proteome 75(12):3720–3732
Yin B et al (2015) Glycoengineering of Chinese hamster ovary cells for enhanced erythropoietin N-glycan branching and sialylation. Biotechnol Bioeng 112(11):2343–2351
Bierhuizen MF, Fukuda M (1992) Expression cloning of cDNA encoding UDP-GlcNAc:Gal beta 1-3-GalNAc-R (GlcNAc to GalNAc) beta 1-6GlcNAc transferase by gene transfer into CHO cells expressing polyoma large tumor antigen. Proc Natl Acad Sci U S A 89(19):9326–9330
Prati EG et al (2000) Engineering of coordinated up- and down-regulation of two glycosyltransferases of the O-glycosylation pathway in Chinese hamster ovary (CHO) cells. Biotechnol Bioeng 68(3):239–244
Minch SL, Kallio PT, Bailey JE (1995) Tissue plasminogen activator coexpressed in Chinese hamster ovary cells with alpha(2,6)-sialyltransferase contains NeuAc alpha(2,6)Gal beta(1,4)Glc-N-AcR linkages. Biotechnol Prog 11(3):348–351
Krzewinski-Recchi MA et al (2003) Identification and functional expression of a second human beta-galactoside alpha2,6-sialyltransferase, ST6Gal II. Eur J Biochem 270(5):950–961
Lee EU, Roth J, Paulson JC (1989) Alteration of terminal glycosylation sequences on N-linked oligosaccharides of Chinese hamster ovary cells by expression of beta-galactoside alpha 2,6-sialyltransferase. J Biol Chem 264(23):13848–13855
Damiani R et al (2009) Stable expression of a human-like sialylated recombinant thyrotropin in a Chinese hamster ovary cell line expressing alpha2,6-sialyltransferase. Protein Expr Purif 67(1):7–14
Weikert S et al (1999) Engineering Chinese hamster ovary cells to maximize sialic acid content of recombinant glycoproteins. Nat Biotechnol 17(11):1116–1121
Raymond C et al (2015) Production of IgGs with a human-like sialylation in CHO cells. BMC Proc 9(Suppl 9):O3
Jeong YT et al (2008) Enhanced sialylation of recombinant erythropoietin in CHO cells by human glycosyltransferase expression. J Microbiol Biotechnol 18(12):1945–1952
Wong NS, Yap MG, Wang DI (2006) Enhancing recombinant glycoprotein sialylation through CMP-sialic acid transporter over expression in Chinese hamster ovary cells. Biotechnol Bioeng 93(5):1005–1016
Jeong YT et al (2009) Enhanced sialylation of recombinant erythropoietin in genetically engineered Chinese-hamster ovary cells. Biotechnol Appl Biochem 52(Pt 4):283–291
Son YD et al (2011) Enhanced sialylation of recombinant human erythropoietin in Chinese hamster ovary cells by combinatorial engineering of selected genes. Glycobiology 21(8):1019–1028
Burg M, Muthing J (2001) Characterization of cytosolic sialidase from Chinese hamster ovary cells: part I: cloning and expression of soluble sialidase in Escherichia coli. Carbohydr Res 330(3):335–346
Ngantung FA et al (2006) RNA interference of sialidase improves glycoprotein sialic acid content consistency. Biotechnol Bioeng 95(1):106–119
Munzert E et al (1997) Production of recombinant human antithrombin III on 20-L bioreactor scale: correlation of supernatant neuraminidase activity, desialylation, and decrease of biological activity of recombinant glycoprotein. Biotechnol Bioeng 56(4):441–448
Ferrari J et al (1998) Chinese hamster ovary cells with constitutively expressed sialidase antisense RNA produce recombinant DNase in batch culture with increased sialic acid. Biotechnol Bioeng 60(5):589–595
Van Dyk DD et al (2003) Identification of cellular changes associated with increased production of human growth hormone in a recombinant Chinese hamster ovary cell line. Proteomics 3(2):147–156
Zhang M et al (2010) Enhancing glycoprotein sialylation by targeted gene silencing in mammalian cells. Biotechnol Bioeng 105(6):1094–1105
Tousi F, Hancock WS, Hincapie M (2011) Technologies and strategies for glycoproteomics and glycomics and their application to clinical biomarker research. Anal Methods 3(1):20–32
Zhang Y, Yin H, Lu H (2012) Recent progress in quantitative glycoproteomics. Glycoconj J 29(5–6):249–258
Ito S, Hayama K, Hirabayashi J (2009) Enrichment strategies for glycopeptides. Methods Mol Biol 534:195–203
Hua S, An HJ (2012) Glycoscience aids in biomarker discovery. BMB Rep 45(6):323–330
Furukawa J-i, Fujitani N, Shinohara Y (2013) Recent advances in cellular glycomic analyses. Biomol Ther 3(1):198
Bennun SV et al (2013) Integration of the transcriptome and glycome for identification of glycan cell signatures. PLoS Comput Biol 9(1):e1002813
Ranzinger R et al (2011) GlycomeDB—a unified database for carbohydrate structures. Nucleic Acids Res 39(Database issue):D373–D376
von der Lieth CW et al (2011) EUROCarbDB: an open-access platform for glycoinformatics. Glycobiology 21(4):493–502
Akune Y et al (2010) The RINGS resource for glycome informatics analysis and data mining on the web. OMICS 14(4):475–486
Lutteke T et al (2006) GLYCOSCIENCES.de: an Internet portal to support glycomics and glycobiology research. Glycobiology 16(5):71R–81R
Krambeck FJ et al (2009) A mathematical model to derive N-glycan structures and cellular enzyme activities from mass spectrometric data. Glycobiology 19(11):1163–1175
Yang S, Zhang H (2014) Glycomic analysis of glycans released from glycoproteins using chemical immobilization and mass spectrometry. Curr Protoc Chem Biol 6(3):191–208
Tian Y, Zhang H (2010) Glycoproteomics and clinical applications. Proteomics Clin Appl 4(2):124–132
Megger DA et al (2013) Label-free quantification in clinical proteomics. Biochim Biophys Acta 1834(8):1581–1590
Kashyap MK et al (2010) SILAC-based quantitative proteomic approach to identify potential biomarkers from the esophageal squamous cell carcinoma secretome. Cancer Biol Ther 10(8):796–810
Tian Y, Bova GS, Zhang H (2011) Quantitative glycoproteomic analysis of optimal cutting temperature-embedded frozen tissues identifying glycoproteins associated with aggressive prostate cancer. Anal Chem 83(18):7013–7019
Raso C et al (2012) Characterization of breast cancer interstitial fluids by TmT labeling, LTQ-Orbitrap Velos mass spectrometry, and pathway analysis. J Proteome Res 11(6):3199–3210
Zhang H et al (2003) Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol 21(6):660–666
Baycin-Hizal D et al (2012) Proteomic analysis of Chinese hamster ovary cells. J Proteome Res 11(11):5265–5276
Yang S et al (2015) QUANTITY: an isobaric tag for quantitative glycomics. Sci Rep 5:17585
Sun S et al (2016) Comprehensive analysis of protein glycosylation by solid-phase extraction of N-linked glycans and glycosite-containing peptides. Nat Biotechnol 34(1):84–88
Toghi Eshghi S et al (2015) GPQuest: a spectral library matching algorithm for site-specific assignment of tandem mass spectra to intact N-glycopeptides. Anal Chem 87(10):5181–5188
Furukawa J et al (2015) Quantitative O-glycomics by microwave-assisted beta-elimination in the presence of pyrazolone analogues. Anal Chem 87(15):7524–7528
Lauc G et al (2010) Genomics meets glycomics—the first GWAS study of human N-glycome identifies HNF1α as a master regulator of plasma protein fucosylation. PLoS Genet 6(12):e1001256
Huffman JE et al (2011) Polymorphisms in B3GAT1, SLC9A9 and MGAT5 are associated with variation within the human plasma N-glycome of 3533 European adults. Hum Mol Genet 20(24):5000–5011
Zoldos V et al (2012) Epigenetic silencing of HNF1A associates with changes in the composition of the human plasma N-glycome. Epigenetics 7(2):164–172
Saldova R et al (2011) 5-AZA-2′-deoxycytidine induced demethylation influences N-glycosylation of secreted glycoproteins in ovarian cancer. Epigenetics 6(11):1362–1372
Nairn AV et al (2012) Regulation of glycan structures in murine embryonic stem cells: combined transcript profiling of glycan-related genes and glycan structural analysis. J Biol Chem 287(45):37835–37856
Tan Z et al (2014) Altered N-glycan expression profile in epithelial-to-mesenchymal transition of NMuMG cells revealed by an integrated strategy using mass spectrometry and glycogene and lectin microarray analysis. J Proteome Res 13(6):2783–2795
Yang Z et al (2015) Engineered CHO cells for production of diverse, homogeneous glycoproteins. Nat Biotechnol 33(8):842–844
Chenu S et al (2003) Reduction of CMP-N-acetylneuraminic acid hydroxylase activity in engineered Chinese hamster ovary cells using an antisense-RNA strategy. Biochim Biophys Acta 1622(2):133–144
Zhang X, Lok SH, Kon OL (1998) Stable expression of human alpha-2,6-sialyltransferase in Chinese hamster ovary cells: functional consequences for human erythropoietin expression and bioactivity. Biochim Biophys Acta 1425(3):441–452
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG
About this chapter
Cite this chapter
Heffner, K.M., Wang, Q., Hizal, D.B., Can, Ö., Betenbaugh, M.J. (2018). Glycoengineering of Mammalian Expression Systems on a Cellular Level. In: Rapp, E., Reichl, U. (eds) Advances in Glycobiotechnology. Advances in Biochemical Engineering/Biotechnology, vol 175. Springer, Cham. https://doi.org/10.1007/10_2017_57
Download citation
DOI: https://doi.org/10.1007/10_2017_57
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-69589-7
Online ISBN: 978-3-030-69590-3
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)