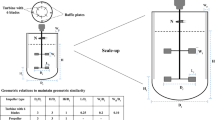
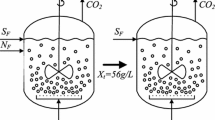
Abstract
This chapter addresses the update progress in bioprocess engineering. In addition to an overview of the theory of multi-scale analysis for fermentation process, examples of scale-up practice combining microbial physiological parameters with bioreactor fluid dynamics are also described. Furthermore, the methodology for process optimization and bioreactor scale-up by integrating fluid dynamics with biokinetics is highlighted. In addition to a short review of the heterogeneous environment in large-scale bioreactor and its effect, a scale-down strategy for investigating this issue is addressed. Mathematical models and simulation methodology for integrating flow field in the reactor and microbial kinetics response are described. Finally, a comprehensive discussion on the advantages and challenges of the model-driven scale-up method is given at the end of this chapter.
Graphical Abstract

Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Bioprocess engineering
- Scale-up
- Bioprocess monitoring
- Fluid dynamics
- Microbial kinetics
- Mathematical model
1 Introduction
With the ever-growing demand for foods, nutriceuticals, pharmaceuticals, fuels, and materials as well as for sustainable development of economy and environment, microbial fermentations using low-cost and renewable feedstocks have become increasingly important. Great effort has been endeavored for improving yields, titers, and productivities of aimed products through bioprocess engineering strategies since the very early beginning of the fermentation industry. Among them, optimization and scale-up method toward industrial process shows great importance. In principle, fermentation capacity is of great extent depending on two compelling parts, i.e., the cellular machinery (determined by gene functions and individual enzyme kinetics) and the extracellular environment (determined by fluid dynamics in bioreactor) [1]. In this regard, it is a permanent task and challenge for biotechnological research to discover and gain more knowledge of cell kinetics and bioreactor fluid dynamics, and interaction between these two parts, in order to accelerate the transition process from laboratory investigation to industrial application [2].
In the past four decades, a huge amount of advanced process monitoring techniques have been developed and applied for bioprocess monitoring and control, e.g., IR spectroscopy for online real-time measurement of glucose, glutamate, fructose, glutamine, proline, and ammonia [3]; Raman spectroscopy for measurement of glucose, acetate, formate, lactate and phenylalanine, and carotenoid production [4]; capacitance sensor for biomass measurement [5]; and online MS for measurement of real-time concentration of O2 and CO2 in exhaust gas [6]. These rich real-time data, together with the increasing mega data along with the advent of OMICS techniques, make tens of thousands of parameters can be monitored and analyzed simultaneously [7, 8] at different scales. However, if a certain phenomenon happening on one scale is regarded as the main research objective, it is hard to discover the relevance among different scales because of difficulties with micro- and macro-statistical processing of the data. Thus the multi-scale analysis methodology on bioprocess becomes an issue of interest [9].
Another important issue of bioprocess scale-up is heterogeneous flow field in the industrial-scale bioreactor and its negative effect on the cell physiology [10, 11]. Bioprocesses conducted in large-scale bioreactors always face mixing or mass transfer problem which may not occur in laboratory- or bench-scale bioreactors. It has been observed that biomass yield of E. coli decreased by 20 % in large-scale bioreactor (12 m3) compared to in bench-scale bioreactor [12]. Heterogeneous environment in the large-scale bioreactor is deemed to main cause of the scale-up issue; thus, experimental investigation using scale-down system in laboratory scale was proposed to gain more insights on this issue. Neubauer [13] reviewed different scale-down simulation systems and concluded that the scale-down system provided improved possibilities to evaluate how a bioprocess would behave in the final industrial scale. However, lack of quantitative information of flow field both in large-scale bioreactor and scale-down system makes it hard to determine whether they are under the same heterogeneity. Computational fluid dynamics (CFD) tools can possibly be the best solution to this problem, but few researches have been reported regarding this. Using the multi-scale analysis technique, more information about the cell physiology under different scales can be gained. Combined with flow field information in different scale bioreactors by CFD, it can be used to rationally direct the bioprocess scale-up [14]. To some extent, this can solve the real industrial problem quickly without deep insight into the cell kinetic mechanism.
Mathematical models have been used to understand, predict, and optimize the properties and behavior of cells and the bioprocess [15]. Kinetic models can be used for different purposes, e.g., enhancing substrate utilization and product yield, detecting metabolic engineering target, and improving process design [16]. For rational scale-up of bioprocess, kinetic models that predict cell behavior in dynamic external environments coupled with fluid dynamics models that describe mass transfer and mixing in the bioreactor can be employed to predict output of the whole culture system under different scales. This issue was raised by Vrabel et al. [17] and Schamalzriedt et al. In their research using Euler–Euler simulation framework, however, historical effects of external environment on cells were not considered. Lapin [18, 19] solved this problem by using Euler–Lagrange simulation framework, which simulated the biophase using Lagrangian frame of reference. However, much more precise and fast dynamic response experiments on cells should be designed to validate the proposed models.
In this chapter, we discuss mainly the advances and applications of bioprocess techniques in the bioindustrial field. New improvement of the multi-scale analysis of fermentation process and its successful practices in industrial bioprocess were addressed. Cause of heterogeneity in large-scale bioreactor and its effect on microbial physiology will be analyzed first, and then, experimental scale-down investigation method and mathematical modeling are reviewed to decipher this issue. The development and new perspective of bioprocess engineering are also proposed at the end of this chapter.
2 Bioprocess Research Based on Multi-scale Analysis of Fermentation Process
2.1 Theory of Multi-scale Analysis for Fermentation Process
Nowadays, to our knowledge, strain improvement strategy relies stepwise on an oriented genome-scale restructure, rather than random mutagenesis. Following the acquired high-yield strain, how to effectively and efficiently apply it and scale-up to industrial-scale challenges both academic and industrial researchers. Traditional scale-up is mainly based on the principle of similarity and dimensional analysis, such as using the same specific power consumption, KLa, impeller speed, and mixing time as scale-up criteria. Under this circumstance, however, no biological properties are taken into account, and thus, no universal principle can be deduced for successful scale-up [20]. In other words, biologically fermentation system is a complex system with a large number of elements, building blocks, or agents, capable of buffering stimuli with one another and with their environments [21]. To address this problem, in the wake of a long-term accumulation of theory and practice, the multi-scale analysis method dealing with the complex interplay between the cellular machinery and the extracellular environment was at the first time proposed by Zhang et al. [22] and has already successfully found its applicability in the production of varying commercial products, such as penicillin, erythromycin, chlortetracycline, inosine, guanosine, recombinant human serum, and malaria vaccine.
It should be noted that output of real fermentation process is limited by strain capacity but driven by nutrients environment that surrounding microbial cells, especially in industrial-scale bioreactor. For instance, it is necessary to maintain the dissolved oxygen (DO) tension above 30 % during penicillin fermentation; otherwise, the penicillin synthesis is irreversibly affected [23]. While, in other cases, oxygen limitation strategy is very helpful, e.g., during glucoamylase fermentation using Aspergillus niger [24]. Most interesting is the fact that there are some robust strains and wild-type or engineered strains, less sensitive to environmental changes in a certain range, which to a great extent relieves the burden of process optimization and scale-up [25, 26]. In most cases, however, microbial physiology and product formation are prone to be influenced by changes in the extracellular environment. Nowadays, there is an increasing tendency of studying effects of environmental gradients on metabolism, flux, and growth rate [25, 27–29]. Due to insufficient mixing and mass transfer limitations in large-scale fermentors, various gradients, including substrate gradient, DO gradient, pH gradient, and carbon dioxide gradient, do occur inevitably and the cells are always experiencing feast/famine cycles. As a consequence, biomass, productivity, and yield are severely affected [30]. As first proposed by Oosterhuis, the scale-down simulation experiment has been employed to investigate local limitation effects in the large-scale fermentors, accordingly. Since then, a couple of scale-down simulation systems have been designed, which could accelerate the prediction of large-scale performance [13, 20, 31, 32]. It is, therefore, of great significance to detect environmental changes in the course of fermentation and adjust corresponding regimes, such as feeding strategy, to enhance productivity and yield, afterward [33].
Aided by numerous instruments mounted on the fermentor, the multi-scale method using many online critical parameters, such as DO, pH, temperature (T), oxygen uptake rate (OUR), carbon dioxide emission rate (CER), and respiratory quotient (RQ), depicts a holistic picture of fermentation process. In other words, dynamic tendency of bioprocess is very important for getting a deeper insight into microscopic metabolism of the culture over the entire process. For example, biomass is one of the most important physiological parameters in fermentation process, which is usually expressed as dry cell weight (DCW). However, DCW, the aggregation of both active and inactive cells and hence, fails to reflect real-time microbial physiology. Colony-forming units (CFU) is thus applied to estimate the viable cells, but it is time consuming and difficult to know what happens in a fermentor as a function of time. Thus, online capacitance probe was used to get accurate determination of viable cells and reflects time-course of active biomass, effectively and efficiently [5]. With increasing amount of biosensors being available for detecting changes in a real bioprocess, the multi-scale analysis method using the correlation and tendency of process parameters will be a more powerful methodology in studying practices and developing optimized strategies.
2.2 Process Optimization in the Context of Association Analysis of Process Parameters
The multi-scale analysis method describing the highly correlated process parameters has been employed to analyze dynamics of the culture. On one hand, biosensors mounted in the fermentor can read direct parameters such as DO, T, stirrer speed, and gas flow rate; on the other hand, indirect parameters standing for time-course physiology and metabolism of the strain such as OUR, CER, and RQ can be on line calculated with the data acquisition software package. A number of successful cases relating to bioprocess scale-up were realized by the multi-scale analysis method.
As a case in point, in the late-phase production of guanosine, a typical phenomenon was that OUR, CER, productivity, and yield of guanosine were rapidly decreasing, while sugar and ammonia consumption were still increasing. Considering the carbon balance, it was deduced that some intermediates of metabolism, such as amino acids, organic acids, or some other nitrogenous substances, had accumulated, and the metabolic flux had shifted to synthesis of these intermediates, other than guanosine. As evidenced by the time-course of by-products, key enzymatic activities, and stoichiometric calculation of metabolic flux shift, it was concluded that late-phase production of guanosine competed against alanine accumulation, which provided possible direction for metabolic engineering and also for process optimization [6, 34, 35]. Another successful case was the optimization of recombinant human serum albumin by an engineered P. pastoris. Generally, a severe phenomenon was the emergence of zero dissolved oxygen level after fermentation for 36 h, which was the main problem to be solved. A novel medium and feeding strategy were proposed to address this problem by analysis of online parameters using the multi-scale analysis method [9].
In parallel, the multi-scale analysis method was also successfully applied to optimization of the production of secondary metabolites, such as erythromycin, penicillin, avermectin, and tylosin. Penicillin is the first bio-based pharmaceutical discovered by Alexander Fleming, and its production titer nowadays reaches over 100,000 times higher than the original strain [36]. However, from stoichiometric perspective, the theoretical value was calculated to be 0.50 mol/mol glucose, where of course, direct sulfhydrylation was only considered and no by-product was taken into account [37]. Therefore, it is still a long way to optimize the strain and process strategy. For example, on a large-scale penicillin production, one of the most important regulation strategies is breakup of foam by frequent addition of antifoam reagent, soybean oil. Soybean oil in fact has other mechanisms of function because its addition brings either positive or negative effects if not properly designed. To this end, the multi-scale analysis method was used to analyze the underlying mechanism of addition of soybean oil. In conclusion, soybean oil may be: (a) used as carbon source. One mole of oil is equivalent to more than two moles of glucose; hence, the addition of a large amount of oil will result in abundant carbon source, which in turn influence the specific growth rate and thus impair final productivity. (b) correlated with oxygen supply. An appropriate amount of foam is in favor of oxygen transfer especially in early stage of fermentation. Moreover, another effect of soybean oil drives carbon dioxide out of foam and reduces its side effect. As a consequence, it is suggested that soybean oil should be intermittently added in small amounts.
Derived from the multi-scale analysis method, “chaotic phenomenon” was frequently used for characterization of secondary metabolism. What is the “chaotic phenomenon”? After years’ study of synthesis of secondary metabolites, e.g., antibiotics, a general phenomenon is frequently happening in both laboratory and industry cases that the initial fermentation phase shows process polymorphism and uncertainty, and tiny difference of initial input may exert a huge impact on the final result. For instance, the “chaotic phenomenon” did occur in the production of erythromycin when different carbon sources were used. It was drawn from the data-trend curves that soybean meal, instead of glucose, via amino acid metabolism might function in supplying erythromycin with the carbon skeleton. Properly controlled, the productivity and yield of erythromycin would be noticeably enhanced [38]. In parallel with discovery of the “chaotic phenomenon,” stringent response, a common metabolic regulation mechanism found in a wide range of prokaryotes and also in plants is involved in the synthesis and accumulation of guanosine tetra-(ppGpp) or pentaphosphates (pppGpp) when cells encounter nutrient starvation. This phenomenon, a global transcriptional regulation response, has considerable functions in growth rate control, DNA maintenance, protein turnover, sporulation, and also in antibiotic synthesis [39–42]. Therefore, the multi-scale analysis method accelerates profound comprehension of the bioprocess and may also provide clues for explanation of some underlying mechanisms involving the molecular scale (genetics), the cellular scale (metabolic regulation), and the reactor scale (process control). It can be seen that the multi-scale analysis method provides series of principles for investigating the fermentation process, which can be seen as a complex system [43].
2.3 Successful Scale-up by Combination of Process Parameters and Fluid Dynamics
Two important issues, namely physiology of microbes and flow field in fermentors, are highly interconnected during the whole fermentation process and affect final fermentation performance. Of great importance is to understand relationship between these two aspects, which will further accelerate the scale-up process [44]. Following the multi-scale analysis method describing real-time physiology of cells, a huge amount of efforts have been taken to understand flow field of fermentation system. CFD has been established as a very useful tool in solving serial problems such as flow of fluid, mixing, transfer, and chemical reactions [45, 46]. CFD technology follows the fundamentals where the conservation of fluid mass, momentum, and energy are governed, and it has been used to find out the bottleneck involved in industrial scale-up.
As a case in point, on a 12 m3 cephalosporin C (CPC) production scale, Yang et al. simulated two different impeller combinations using CFD, in which one was the conventional radial impellers and the other was the combination of radial and axial impellers. From the point of simulation results, it was conceivable that the novel impeller configuration generated more homogeneous conditions. In industrial practice, soybean oil is widely used as carbon source in the antibiotics fermentation process. Though it is poorly soluble in broth, soybean oil is normally fed at the top of fermentor, which results in oil gradient in case of poor mixing. The discrepancy of process parameter RQ value under these two impeller configurations reveals different consumption rate of soybean oil. It was found that RQ profiles branched after the first 50 h fermentation between the two impeller combinations, and RQ value under novel impeller combination was much closer to the theoretical value. This indicates that more homogeneous soybean oil concentration was formed under novel impeller combination due to its more effective mixing capacity. Moreover, hydrodynamic environment generated by the novel combination may favor formation of dispersed arthrospores, rather than mycelia, and thus enhance CPC production [47, 48].
In 132 m3 erythromycin fermentation, by using CFD simulation of fluid dynamics on different scales, it was concluded that the main cause of impairment of physiological metabolism and erythromycin formation was the decrease of OTR as volume increased. The OUR in 132-m3 fermentor was obviously lower than that in 50-L fermentor, which further confirmed the insufficient oxygen supply in the larger-scale bioreactor. Therefore, CFD helps to understand the relationship between flow field in fermentors and physiology of microbes [14]. In conclusion, integration of fluid dynamics and process parameters has been proven to be an efficacious way to comprehensively understand what is going on in different scale fermentors [44].
3 Optimization and Scale-up of Bioprocess Based on Integration of Fluid Dynamics and Biokinetics
3.1 Heterogeneity Environment in Large-scale Bioreactor and Its Impact on Cells
A dynamic environment with large spatial and temporal heterogeneities is always produced in large-scale fermentor especially under fed-batch mode [49]. Several aspects will change inevitably during the scale-up from bench- to industrial-scale [31]. For example, with increasing liquid height, hydrostatic pressure at bottom of the large reactor can easily reach over 1 bar, which in turn results in higher oxygen solubility, and vertical oxygen concentration gradients will then be formed. Mixing time can also easily reach over 2 min in large-scale fermentor up to 19 m3 [50], while it is only in the order of seconds in laboratory-scale bioreactor. Furthermore, pH gradient and temperature gradient are also found in large-scale bioreactors. As Lara et al. [10] has analyzed, when mixing time in the large scale was in the same order of magnitude as the reaction time, heterogeneities can be anticipated. However, it is hard to maintain constant mixing time at different scales because of limitation of power input in the large-scale bioreactor.
It should be noted that cells are alive and highly responsive compared to chemical reactant or catalyst. These above-mentioned heterogeneities can induce multiple physiological responses; different cells have different responses according to their genetic or metabolic control mechanism. As bioreactor scale increases, mixing time may exceed over the bioreaction characteristic time, which, however, is independent of bioreactor size [10, 49]. Thus, cells in large-scale bioreactors may probably experience an oscillating environment which leads to a heterogenetic cell population. This usually causes lower yields and productivities and an increased by-product formation compared to laboratory-scale bioreactors [11, 12, 29]. Therefore, lots of time and effort are taken to study scale-up process, which is a great hurdle for rapid development of bioprocesses from micro-liter cultures to the industrial scale [51]. Understanding the heterogeneity in industrial-scale bioreactors and the performance of cells exposed to an alternating environment is of great importance for efficient enhancement of biotechnical processes.
3.2 Development of Scale-down System and Its Applications
The scale-down concept was first proposed by Kossens and coworkers [52]. Regime analysis was used to interpret the large-scale process by comparing characteristic time of different processes, and scale-down simulation system was then established based on this information [53]. It is proven to be an effective way for investigating the effect of oscillating environments on cell metabolic and physiological behavior in a laboratory-scale equipment. Various scale-down systems have been designed to investigate effects of heterogeneities such as pH, temperature, dissolved oxygen, and substrate on physiology of bacteria [54], yeast [55], filamentous fungi [56], and mammalian cells [57].
Among all kinds of scale-down systems mimicking environmental gradients, the most frequently used can be roughly classified into two groups, namely one-compartment systems and two-compartment systems. Usually, a one-compartment system consists of a stirred tank reactor (STR) [58] or specially designed tubular reactor [59]. A one-compartment scale-down system was designed to simulate dissolved oxygen tension gradient by León-Rodríguez et al. [60]. The results showed that one-compartment system was able to generate sustained oscillating dissolved oxygen profiles. One-compartment system is simple in structure and relies on control strategy to generate gradients. However, in one-compartment scale-down system, all cells are in the same condition which means such approach is unable to mimic the cell population heterogeneity in real bioreactor. While, in a two-compartment system, cells circulate through the two connected compartments, each of which implements a certain different condition. The two-compartment scale-down system can be constructed by either connecting two STRs or connecting one STR with a plug-flow reactor (PFR), which can be used to simulate real cycling times and environmental gradients. The combination of two compartments makes the system more facile to reflect the real large-scale bioreactor. Junne et al. [61] constructed a two-compartment bioreactor with commercially available parts to investigate a non-sporulating B. subtilis response to oscillating glucose and dissolved oxygen concentration and found that carbon flux at excess glucose and low DO concentration was shifted toward ethanol formation, as a result, diminished glucose uptake and altered amino acid synthesis were observed. Sandoval-Basurto et al. [62] implemented a two compartment scale-down system to investigate dissolved oxygen gradient effect on recombinant E. coli. Two STRs with different DO levels were connected. It was found that acetic, lactic, formic, and succinic acids accumulated under oscillating DO conditions.
For scale-down system, it is very important to make sure that it can represent the situation that cells encounter in large-scale bioreactor. Otherwise, the investigation of different oscillating conditions may not be relevant to the key hurdle of the scale-up. To this end, CFD investigation on the large-scale reactor can pave the way for rational design of the scale-down system, as it can provide more detailed and local information about hydrodynamics and mixing. However, the real fluctuating environment in the large-scale bioreactor is a result of both fluid mixing and metabolic response of cells in the reactor. This means not only the fluid dynamics in the reactor, but also kinetic information of the cells should be considered for this purpose. Therefore, an idea of coupling these two aspects comes out for better understanding of the scale-up process.
3.3 Model-driven Rational Scale-up of Bioprocess
Mathematical models describing both fluid dynamics in the reactor and bioreaction kinetics will help to shed bright light on the behavior of the industrial-scale fermentation system, which in the end leads to a rational design of an industrial fermentation process. Since 1980s, this method has been used to simulate the performance of large-scale fermentation system by coupling fluid dynamics model with simple substrate uptake model [63]. However, the complexity of both models makes it more difficult to be conducted in engineering practice; hence, simplified fluid dynamics model of compartment model approach and Monod-type kinetic model were always used at that time [17]. With rapid development of computer hardware and simulation algorithm, nowadays, the CFD software has been used to solve flow field problems up to 120-m3 scale reactor [14]. Integrated models that couple fluid dynamics and microbial kinetics have been used to simulate hydrodynamic effects on filamentous morphology [64] and physiological response [65] in different kinds of reactors.
All the above-mentioned simulation work are based on Euler–Euler approach in which gas, liquid, and biophase are all considered as a continuum and described in terms of their volume fractions. In the past two decades, this approach has been applied to various biological processes [1, 30, 66]. A simple multi-scale kinetic model based on Herbert’s concept was coupled with a CFD model to investigate influence of mixing mechanism in a 1.5-L bioreactor on ethanol fermentation. Compared to experiment results, simulated data showed approximately 5 % error for yield and 14 % error for productivity [67]. Elqotbi successfully simulated the whole 60 h fermentation process of A. niger with limited computational efforts based on the separated solution of the flow field, the mass transfer, and microbial reaction. Such stepwise solution strategy divides the whole fermentation process into three steps—firstly solving the fluid flow field, secondly imposing oxygen mass transfer and bioreaction, and finally updating flow field after bioreaction. This strategy successfully solved the problem of different timescales between hydrodynamics and bioreaction [68]. Segregated model for the biophase using population balance model (PBM) is also coupled to CFD model to address bioreactor-scale effect on the cell population heterogeneity in a recent published work by Morchain et al. [69], which is also based on Euler–Euler frame method.
In fact, the Euler–Euler approach leads to loss of realism if individual history of cells becomes the focus of attention, e.g., when considering cumulative starvation effects in cells during fed-batch fermentation [18]. In contrast, the Euler–Lagrange approach overcomes this problem, in which the fluid phase is treated as a continuum whereas the dispersed biophase is solved by tracking a large number of particles through the calculated flow field [70]. By using this approach, the analysis of lifeline of individual cell in space and time is possible. Lapin et al. [18] first employed this approach to couple yeast glycolytic oscillation with turbulent flow in a 68-L reactor to demonstrate the influence of bioreactor mixing intensity to the synchronization of yeast glycolytic oscillation in a population level. Their results showed that non-ideal mixing condition (N imp = 55 rpm) resulted in slightly diminished degree of synchrony as compared to ideally mixing case (N imp = 165 rpm). They used this approach again to simulate fed-batch fermentation of E. coli in a 900-L bioreactor [19]. In this model, a sugar uptake kinetic model (phosphotransferase system, PTS) was coupled to turbulent flow in bioreactor. The activity of the sugar uptake system depends on the local concentration of glucose as well as the ratio of the intracellular concentrations of phosphoenolpyruvate and pyruvate, which in turn is a function of the history of individual cell. Heterogeneity on the specific sugar uptake rate was observed among the E. coli population. Much of their results were only indirectly verified by experimental observations, but the proposed simulation framework in their work was to some extent a promising method for better understanding the scale-up problem.
To be applied for rational design and scale-up of bioprocess, the integration simulation approach coupling fluid dynamics in bioreactor and cellular kinetics based on both the Euler–Euler and the Euler–Lagrange shows attractive potential. But, there is still a long way to go. One main challenge of this method is how to keep balance of computational expense and simulation accuracy. Another problem of this method is validation of simulation results, even though there are some reports on measurement of intracellular metabolites in single cell level [71], it is really a hard work to get sufficient validation data in even laboratory-scale bioreactor at the present state.
4 Conclusions
Despite the central role of scale-up issue in biotechnology and the large body of literatures, there seems to be no common, universally applicable strategy [72]. It has been ever stated that scale-up is an art rather than an exact science [73]. Indeed, the fermentation process conducted in bioreactor is really a complex system, as the cell, which is alive, has a precise control mechanism which shows different responses to environment perturbations on different scales. That is believed to be the main cause of various scale-up problems.
With the great increase of knowledge of the interplay between cell physiological response and extracellular nutrient conditions, we are approaching more rational scaling up of the bioprocess. To identify process-specific stress factors and to understand the physiological responses to the vessel specific physical conditions, the mutual influences and interactions of the various physical and physiological parameters need to be analyzed in detail [72]. Multi-scale fermentation analytical method coupled with fluid dynamics investigation can promisingly implement this goal and has been applied to optimize and scale-up of different fermentation process, which was proved to be an efficient approach. A holistic scale-up strategy consists of a comprehensive and detailed process characterization of both laboratory-scale and industrial-scale fermentor to identify key process parameters influencing product yield and productivity.
In parallel, with development of both metabolic engineering and systems biotechnology, more and more mathematical models describing cell metabolism and its regulation mechanism have been proposed. But most of them focus on the stoichiometry relations but not kinetic effects. As Almquist et al. [16] pointed out, kinetic model should be a powerful tool for better understanding the metabolic mechanism for their response to either genetic manipulation or environment fluctuations. While it is a challenge to build a holistic realistic kinetic model as little is known about the in vivo mechanisms of enzymes and transporters. Short-term perturbation experiment based on fast sampling technique and precise measurement of intracellular metabolites is a well-performed tool for establishing such in vivo kinetic model. Such model with careful validation can be used to couple CFD model in different scale reactors for rational scale-up of different bioprocesses.
References
Schamalzriedt S et al (2003) Integration of physiology and fluid dynamics. In: Scheper T (ed) Advances in biochemical engineering/biotechnology. Springer, Berlin, pp 19–68
Babel W, Brinkmann U, Müller R (1993) The auxiliary substrate concept—an approach for overcoming limits of microbial performances. Acta Biotechnol 13(3):211–242
Landgrebe D et al (2010) On-line infrared spectroscopy for bioprocess monitoring. Appl Microbiol Biotechnol 88(1):11–22
Lee HLT et al (2004) In situ bioprocess monitoring of Escherichia coli bioreactions using Raman spectroscopy. Vibrational Spectroscopy 35(1–2):131–137
Li L et al (2014) Optimization of polyhydroxyalkanoates fermentations with on-line capacitance measurement. Bioresour Technol 156:216–221
Zhang SL et al (2004) Studies on the multi-scale problems of guanosine fermentation process. Chin J Bioprocess Eng 2(3):23–29
Bonneau R et al (2007) A predictive model for transcriptional control of physiology in a free living cell. Cell 131(7):1354–1356
Ishii N et al (2007) Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science 316:593–597
Zhang SL, Chu J, Zhuang Y (2004) A multi-scale study of industrial fermentation processes and their optimization. Adv Biochem Eng Biotechnol 87:97–150
Lara AR et al (2006) Living with heterogeneities in bioreactors. Mol Biotechnol 34(3):355–381
Enfors S-O et al (2001) Physiological responses to mixing in large scale bioreactors. J Biotechnol 85(2):175–185
Bylund F et al (1998) Substrate gradient formation in the large-scale bioreactor lowers cell yield and increases by-product formation. Bioprocess Eng 18(3):171–180
Neubauer P, Junne S (2010) Scale-down simulators for metabolic analysis of large-scale bioprocesses. Curr Opin Biotechnol 21(1):114–121
Zou X et al (2012) Real-time fluid dynamics investigation and physiological response for erythromycin fermentation scale-up from 50 L to 132 m3 fermenter. Bioprocess Biosyst Eng 35(5):789–800
Tyo KEJ, Kocharin K, Nielsen J (2010) Toward design-based engineering of industrial microbes. Curr Opin Microbiol 13(3):255–262
Almquist J et al (2014) Kinetic models in industrial biotechnology—improving cell factory performance. Metab Eng 24:38–60
Vrabel P et al (2001) CMA: integration of fluid dynamics and microbial kinetics in modelling of large-scale fermentations. Chem Eng J 84(3):463–474
Lapin A, Müller D, Reuss M (2004) Dynamic behavior of microbial populations in stirred bioreactors simulated with euler–lagrange methods: traveling along the lifelines of single cells. Ind Eng Chem Res 43(16):4647–4656
Lapin A, Schmid J, Reuss M (2006) Modeling the dynamics of E. coli populations in the three-dimensional turbulent field of a stirred-tank bioreactor–A structured-segregated approach. Chem Eng Sci 61(14):4783–4797
Wang G et al (2014) Prelude to rational scale-up of penicillin production: a scale-down study. Appl Microbiol Biotechnol 98(6):2359–2369
Ottino JM (2003) Complex systems. AIChE J 49(2):292–299
Zhang SL, Chu J, Zhuang YP (2004) A multi-scale study of industrial fermentation processes and their optimization. In: Biomanufacturing. Springer, Berlin, pp 97–150
Vardar F, Lilly M (1982) Effect of cycling dissolved oxygen concentrations on product formation in penicillin fermentations. Eur J Appl Microbiol Biotechnol 14(4):203–211
Pedersen L et al (2012) Industrial glucoamylase fed-batch benefits from oxygen limitation and high osmolarity. Biotechnol Bioeng 109(1):116–124
Baez A et al (2011) Simulation of dissolved CO2 gradients in a scale-down system: a metabolic and transcriptional study of recombinant Escherichia coli. Biotechnol J 6(8):959–967
Käß F et al (2013) Assessment of robustness against dissolved oxygen/substrate oscillations for C. glutamicum DM1933 in two-compartment bioreactor. Bioprocess Biosyst Eng 1–12
Taymaz-Nikerel H et al (2013) Changes in substrate availability in Escherichia coli lead to rapid metabolite, flux and growth rate responses. Metabol Eng 16:115–129
de Jonge LP et al (2011) Scale-down of penicillin production in Penicillium chrysogenum. Biotechnol J 6(8):944–958
Käß F et al (2014) Process inhomogeneity leads to rapid side product turnover in cultivation of Corynebacterium glutamicum. Microb Cell Fact 13(1):1–11
Larsson G et al (1996) Substrate gradients in bioreactors: origin and consequences. Bioprocess Eng 14(6):281–289
Takors R (2012) Scale-up of microbial processes: impacts, tools and open questions. J Biotechnol 160(1–2):3–9
Noorman H (2011) An industrial perspective on bioreactor scale-down: what we can learn from combined large-scale bioprocess and model fluid studies. Biotechnol J 6(8):934–943
de Jonge L et al (2014) Flux response of glycolysis and storage metabolism during rapid feast/famine conditions in Penicillium chrysogenum using dynamic 13C labeling. Biotechnol J 9(3):372–385
Chu J et al (2005) Correlation between key enzyme activities in the inosine synthetic pathway and inosine production. Process Biochem 40(2):891–894
Chen SX et al (2005) Enhancement of inosine production by Bacillus subtilis through suppression of carbon overflow by sodium citrate. Biotechnol Lett 27(10):689–692
Rokem JS, Lantz AE, Nielsen J (2007) Systems biology of antibiotic production by microorganisms. Nat Prod Rep 24(6):1262–1287
Jorgensen H et al (1995) Metabolic flux distributions in Penicillium chrysogenum during fed-batch cultivations. Biotechnol Bioeng 46(2):117–131
Zhao J et al (2002) Study on chaos of the erythromycin fermentation process. Chin J Antibiot 27(12):717–719
Chatterji D, Kumar Ojha A (2001) Revisiting the stringent response, ppGpp and starvation signaling. Curr Opin Microbiol 4(2):160–165
Kang SG et al (1998) Actinorhodin and undecylprodigiosin production in wild-type and relA mutant strains of Streptomyces coelicolor A3 (2) grown in continuous culture. FEMS Microbiol Lett 168(2):221–226
Hoyt S, Jones GH (1999) relA is required for actinomycin production in Streptomyces antibioticus. J Bacteriol 181(12):3824–3829
Kuhar I, Žgur-Bertok D (1999) Transcription Regulation of the Colicin Kcka Gene Reveals Induction of Colicin Synthesis by Differential Responses to Environmental Signals. J Bacteriol 181(23):7373–7380
Ye X et al (2005) Multi-scale methodology: a key to deciphering systems biology. Front Biosci 10:961–965
Wang Y et al (2009) Industrial bioprocess control and optimization in the context of systems biotechnology. Biotechnol Adv 27(6):989–995
Sharma C, Malhotra D, Rathore AS (2011) Review of computational fluid dynamics applications in biotechnology processes. Biotechnol Prog 27(6):1497–1510
Xia JY et al (2008) Computational investigation of fluid dynamics in a recently developed centrifugal impeller bioreactor. Biochem Eng J 38(3):406–413
Xia JY et al (2009) Fluid dynamics investigation of variant impeller combinations by simulation and fermentation experiment. Biochem Eng J 43(3):252–260
Yang YM et al (2012) A novel impeller configuration to improve fungal physiology performance and energy conservation for cephalosporin C production. J Biotechnol 161(3):250–256
Hewitt CJ, Nienow AW (2007) The scale up of microbial batch and fed-batch fermentation processes. Adv Appl Microbiol 62:105–135
Junker BH (2004) Scale-up methodologies for Escherichia coli and yeast fermentation processes. J Biosci Bioeng 97(6):347–364
Neubauer P et al (2013) Consistent development of bioprocesses from microliter cultures to the industrial scale. Engineering in Life Sciences 13(3):224–238
Oosterhuis NMG et al (1985) Scale-down and optimization studies of the gluconic acid fermentation by Gluconobacter oxydans. Biotechnol Bioeng 27(5):711–720
Sweere APJ, Luyben KCAM, Kossen NWF (1987) Regime analysis and scale-down: tools to investigate the performance of bioreactors. Enzyme Microb Technol 9(7):386–398
Lara AR et al (2006) Transcriptional and metabolic response of recombinant Escherichia coli to spatial dissolved oxygen tension gradients simulated in a scale-down system. Biotechnol Bioeng 93(2):372–385
Lorantfy B, Jazini M, Herwig C (2013) Investigation of the physiological response to oxygen limited process conditions of Pichia pastoris Mut+ strain using a two-compartment scale-down system. J Biosci Bioeng 116(3):371–379
Nasution U et al (2006) Generating short-term kinetic responses of primary metabolism of Penicillium chrysogenum through glucose perturbation in the bioscope mini reactor. Metab Eng 8(5):395–405
Nienow AW et al (2013) Scale-down studies for assessing the impact of different stress parameters on growth and product quality during animal cell culture. Chem Eng Res Des 91(11):2265–2274
Serrato JA et al (2004) Heterogeneous conditions in dissolved oxygen affect N-glycosylation but not productivity of a monoclonal antibody in hybridoma cultures. Biotechnol Bioeng 88(2):176–188
Papagianni M, Mattey M, Kristiansen B (2003) Design of a tubular loop bioreactor for scale-up and scale-down of fermentation processes. Biotechnol Prog 19(5):1498–1504
León-Rodríguez AD, Galindo E, Ramírez OT (2010) Design and characterization of a one-compartment scale-down system for simulating dissolved oxygen tension gradients. J Chem Technol Biotechnol 85(7):950–956
Junne S et al (2011) A two-compartment bioreactor system made of commercial parts for bioprocess scale-down studies: impact of oscillations on Bacillus subtilis fed-batch cultivations. Biotechnol J 6(8):1009–1017
Sandoval-Basurto EA et al (2005) Culture of Escherichia coli under dissolved oxygen gradients simulated in a two-compartment scale-down system: metabolic response and production of recombinant protein. Biotechnol Bioeng 89(4):453–463
Trägårdh C (1988) A hydrodynamic model for the simulation of an aerated agitated fed-batch fermenter. In: BHRA (ed) Proceedings of the Second International Conference on Bioreactor Fluid Dynamics, Cambridge, p 117–131
Yu L et al (2012) Hydrodynamic and kinetic study of cellulase production by Trichoderma reesei with pellet morphology. Biotechnol Bioeng 109(7):1755–1768
Bannari R et al (2012) A model for cellulase production from Trichoderma reesei in an airlift reactor. Biotechnol Bioeng 109(8):2025–2038
Noorman H et al (1993) Measurements and computational fluid dynamics simulation of Saccharomyces cerevisiae production in a 30 m3 stirred tank reactor. In: Proceedings of International Symposium on Bioreactor Performance, Helsingùr, Denmark
Teng ELW, P Kumar, Y Samyudia (2010) Computational fluid dynamics of mixing in aerated bioreactors. In: Proceedings of International Conference on Biology, Environment and Chemistry
Elqotbi M et al (2013) CFD modelling of two-phase stirred bioreaction systems by segregated solution of the Euler-Euler model. Comput Chem Eng 48:113–120
Morchain J, Gabelle J-C, Cockx A (2014) A coupled population balance model and CFD approach for the simulation of mixing issues in lab-scale and industrial bioreactors. AIChE J 60(1):27–40
Lapin A, Klann M, Reuss M (2010) Multi-scale spatio-temporal modeling: lifelines of microorganisms in bioreactors and tracking molecules in cells. In: Wittmann C, Krull R (eds) Biosystems engineering II. Springer, Berlin, pp 23–43
Grünberger A et al (2012) A disposable picolitre bioreactor for cultivation and investigation of industrially relevant bacteria on the single cell level. Lab Chip 12:2060–2068
Schmidt FR (2005) Optimization and scale up of industrial fermentation processes. Appl Microbiol Biotechnol 68:425–435
Humphrey A (1998) Shake flask to fermentor: what have we learned? Biotechnol Prog 14:3–7
Acknowledgment
Financial support of State Key Development Program of Basic Research of China (973 Program, Grant No. 2013CB733600), NWO-MoST Joint Program (2013DFG32630), National High Technology Research and Development Program of China (863 Program, Grant No. 2012AA021201 and 2014AA021701), and China Ministry of Science and Technology under Contract (Grant No. 2012BAI44G00) are gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Xia, J. et al. (2015). Advances and Practices of Bioprocess Scale-up. In: Bao, J., Ye, Q., Zhong, JJ. (eds) Bioreactor Engineering Research and Industrial Applications II. Advances in Biochemical Engineering/Biotechnology, vol 152. Springer, Berlin, Heidelberg. https://doi.org/10.1007/10_2014_293
Download citation
DOI: https://doi.org/10.1007/10_2014_293
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-662-48346-6
Online ISBN: 978-3-662-48347-3
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)