Abstract
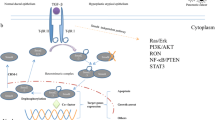
Pancreatic ductal adenocarcinoma (PDAC) is a deadly cancer with a 9% 5-year survival rate. For reasons that are not readily evident, KRAS is mutated in 90–95% of PDAC cases, and this truncal alteration is associated with a high frequency of mutations in crucially important tumor suppressor genes, most notably CDKN2A (~90%), a gene that encodes p16, TP53 (~70%), and SMAD4 (~50%). Concomitantly, there is overexpression of transforming growth factor beta (TGF-β) isoforms and of high-affinity tyrosine kinase receptors (TKRs) and their ligands. Enhanced cancer cell proliferation and migration mediated by TKRs, combined with loss of beneficial TGF-β-dependent pathways required to restrain uncontrolled cell proliferation, contributes to PDAC’s biological aggressiveness. This chapter provides an overview of these issues and focuses on the role of alterations in Smad4 expression and function and aberrant TGF-β signaling that combine to promote pancreatic cancer growth through cell autonomous and paracrine actions, thereby contributing in an important manner to PDAC pathobiology.
Similar content being viewed by others
References
Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2017;67:7–30.
Menke A, Casagrande S, Geiss L, Cowie CC. Prevalence of and trends in diabetes among adults in the United States, 1988–2012. J Am Med Assoc. 2015;314:1021–9.
Aggarwal G, Kamada P, Chari S. Prevalence of diabetes mellitus in pancreatic cancer compared to common cancers. Pancreas. 2013;42:198–201.
Tang H, Dong X, Hassan M, Abbruzzese JL, Li D. Body mass index and obesity- and diabetes-associated genotypes and risk for pancreatic cancer. Cancer Epidemiol Biomark Prev. 2011;20:779–92.
Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–21.
Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371:1039–49.
Paulson AS, Tran Cao HS, Tempero MA, Lowy AM. Therapeutic advances in pancreatic cancer. Gastroenterology. 2013;144:1316–26.
Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20:1218–49.
Preis M, Korc M. Signaling pathways in pancreatic cancer. Crit Rev Eukaryot Gene Expr. 2011;21:115–29.
Provenzano PP, Cuevas C, Chang AE, Goel K, Von Hoff D, Hingorani SR. Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21:418–29.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74.
Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2001;321:1801–6.
Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501.
Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52.
Kingsley DM. The TGF-beta superfamily: new members, new receptors, and new genetic tests of function in different organisms. Genes Dev. 1994;8:133–46.
Bragdon B, Moseychuk O, Saldanha S, King D, Julian J, Nohe A. Bone morphogenetic proteins: a critical review. Cell Signal. 2011;23(4):609–20.
Wu MY, Hill CS. TGF-beta superfamily signaling in embryonic development and homeostasis. Dev Cell. 2009;16:329–43.
Gold L. The role for transforming growth factor-beta (TGF-beta) in human cancer. Clin Rev Oncog. 1999;10:303–60.
Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700.
Weiss A, Attisano L. The TGFbeta superfamily signaling pathway. Rev Dev Biol. 2013;2:47–63.
Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–810.
Tsukazaki T, Chiang TA, Davison AF, Attisano L, Wrana JL. SARA, a FYVE domain protein that recruits Smad2 to the TGFbeta receptor. Cell. 1998;95:779–91.
Feng XH, Derynck R. Specificity and versatility in TGF-beta signaling through Smads. Annu Rev Cell Dev Biol. 2005;21:659–93.
Holtzhausen A, Golzio C, How T, Lee YH, Schiemann WP, Katsanis N, et al. Novel bone morphogenetic protein signaling through Smad2 and Smad3 to regulate cancer progression and development. FASEB J. 2014;28:1248–67.
Nicolás FJ, De Bosscher K, Schmierer B, Hill CS. Analysis of Smad nucleocytoplasmic shuttling in living cells. J Cell Sci. 2004;117:4113–25.
Germain S, Howell M, Esslemont GM, Hill CS. Homeodomain and winged-helix transcription factors recruit activated Smads to distinct promoter elements via a common Smad interaction motif. Genes Dev. 2000;14:435–51.
Hill CS. Transcriptional control by the SMADs. Cold Spring Harb Perspect Biol. 2016;8(10). pii: a022079. doi:10.1101/cshperspect.a022079.
Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–84.
Dai F, Shen T, Li Z, Lin X, Feng XH. PPM1A dephosphorylates RanBP3 to enable efficient nuclear export of Smad2 and Smad3. EMBO Rep. 2011;12:1175–81.
Watanabe M, Masuyama N, Fukuda M, Nishida E. Regulation of intracellular dynamics of Smad4 by its leucine-rich nuclear export signal. EMBO Rep. 2000;1:176–82.
Topper JN, Cai J, Qiu Y, Anderson KR, Xu YY, Deeds JD, et al. Vascular MADs: two novel MAD-related genes selectively inducible by flow in human vascular endothelium. Proc Natl Acad Sci USA. 1997;94:9314–9.
Ebisawa T, Fukuchi M, Murakami G, Chiba T, Tanaka K, Imamura T, et al. Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J Biol Chem. 2001;276:12477–80.
Ogunjimi AA, Briant DJ, Pece-Barbara N, Le Roy C, Di Guglielmo GM, Kavsak P, et al. Regulation of Smurf2 ubiquitin ligase activity by anchoring the E2 to the HECT domain. Mol Cell. 2005;19:297–308.
Eichhorn PJ, Rodon L, Gonzalez-Junca A, Dirac A, Gili M, Martinez-Saez E, et al. USP15 stabilizes TGF-β receptor I and promotes oncogenesis through the activation of TGF-β signaling in glioblastoma. Nat Med. 2012;18:429–35.
Shi W, Sun C, He B, Xiong W, Shi X, Yao D, et al. GADD34-PP1c recruited by Smad7 dephosphorylates TGFbeta type I receptor. J Cell Biol. 2004;164:291–300.
Yan X, Lin Z, Chen F, Zhao X, Chen H, Ning Y, et al. Human BAMBI cooperates with Smad7 to inhibit transforming growth factor-beta signaling. J Biol Chem. 2009;284:30097–104.
Nagano Y, Mavrakis KJ, Lee KL, Fujii T, Koinuma D, Sase H, et al. Arkadia induces degradation of SnoN and c-ski to enhance transforming growth factor-beta signaling. J Biol Chem. 2007;282:20492–501.
Lonn P, van der Heide LP, Dahl M, Hellman U, Heldin CH, Moustakas A. PARP-1 attenuates Smad-mediated transcription. Mol Cell. 2010;40:521–32.
Inoue Y, Itoh Y, Abe K, Okamoto T, Daitoku H, Fukamizu A, et al. Smad3 is acetylated by p300/CBP to regulate its transactivation activity. Oncogene. 2007;26:500–8.
Eifler K, Vertegaal AC. SUMOylation-mediated regulation of cell cycle progression and cancer. Trends Biochem Sci. 2015;40:779–93.
Yamanaka Y, Friess H, Buchler M, Beger HG, Gold LI, Korc M. Synthesis and expression of transforming growth factor beta-1, beta-2, and beta-3 in the endocrine and exocrine pancreas. Diabetes. 1993;42:746–56.
Bottinger EP, Jakubczak JL, Roberts IS, Mumy M, Hemmati P, Bagnall K, et al. Expression of a dominant-negative mutant TGF-beta type II receptor in transgenic mice reveals essential roles for TGF-beta in regulation of growth and differentiation in the exocrine pancreas. EMBO J. 1997;16:2621–33.
Wildi S, Kleeff J, Mayerle J, Zimmermann A, Bottinger EP, Wakefield L, et al. Suppression of transforming growth factor beta signalling aborts caerulein induced pancreatitis and eliminates restricted stimulation at high caerulein concentrations. Gut. 2007;56:685–92.
Chaudhury A, Howe PH. The tale of transforming growth factor-beta (TGFbeta) signaling: a soigné enigma. IUBMB Life. 2009;61:929–39.
Baldwin RL, Korc M. Growth inhibition of human pancreatic carcinoma cells by transforming growth factor beta-1. Growth Factors. 1993;8:23–34.
Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–8.
Grady WM, Myeroff LL, Swinler SE, Rajput A, Thiagalingam S, Lutterbaugh JD, et al. Mutational inactivation of transforming growth factor beta receptor type II in microsatellite stable colon cancers. Cancer Res. 1999;59:320–4.
Goggins M, Shekher M, Turnacioglu K, Yeo CJ, Hruban RH, Kern SE. Genetic alterations of the transforming growth factor beta receptor genes in pancreatic and biliary adenocarcinomas. Cancer Res. 1998;58:5329–32.
Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, Rozenblum E, et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–3.
Xu J, Attisano L. Mutations in the tumor suppressors Smad2 and Smad4 inactivate transforming growth factor beta signaling by targeting Smads to the ubiquitin-proteasome pathway. Proc Natl Acad Sci USA. 2000;97:4820–5.
Kleeff J, Maruyama H, Friess H, Buchler MW, Falb D, Korc M. Smad6 suppresses TGF-beta-induced growth inhibition in COLO-357 pancreatic cancer cells and is overexpressed in pancreatic cancer. Biochem Biophys Res Commun. 1999;255:268–73.
Kleeff J, Ishiwata T, Maruyama H, Friess H, Truong P, Büchler MW, et al. The TGF-beta signaling inhibitor Smad7 enhances tumorigenicity in pancreatic cancer. Oncogene. 1999;18:5363–72.
Schutte M, Hruban RH, Geradts J, Maynard R, Hilgers W, Rabindran SK, et al. Abrogation of the Rb/p16 tumor-suppressive pathway in virtually all pancreatic carcinomas. Cancer Res. 1997;57:3126–30.
Carrière C, Gore AJ, Norris AM, Gunn JR, Young AL, Longnecker DS, et al. Deletion of Rb accelerates pancreatic carcinogenesis by oncogenic Kras and impairs senescence in premalignant lesions. Gastroenterology. 2011;141:1091–101.
Gore AJ, Deitz SL, Palam LR, Craven KE, Korc M. Pancreatic cancer-associated retinoblastoma 1 dysfunction enables TGF-β to promote proliferation. J Clin Invest. 2014;124:338–52.
Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–6.
Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69.
Hirata H, Ueno K, Shahryari V, Tanaka Y, Tabatabai ZL, Hinoda Y, et al. Oncogenic miRNA-182-5p targets Smad4 and RECK in human bladder cancer. PLoS One. 2012;7:e51056. doi:10.1371/journal.pone.0051056.
Zhang Y, Fan KJ, Sun Q, Chen AZ, Shen WL, Zhao ZH, et al. Functional screening for miRNAs targeting Smad4 identified miR-199a as a negative regulator of TGF-β signalling pathway. Nucleic Acids Res. 2012;40:9286–97.
Wang Y, Ren J, Gao Y, Ma JZ, Toh HC, Chow P, et al. MicroRNA-224 targets SMAD family member 4 to promote cell proliferation and negatively influence patient survival. PLoS One. 2013;8(7):e68744. doi:10.1371/journal.pone.0068744.
Hao J, Zhang S, Zhou Y, Liu C, Hu X, Shao C. MicroRNA 421 suppresses DPC4/Smad4 in pancreatic cancer. Biochem Biophys Res Commun. 2011;406:552–7.
Hao J, Zhang S, Zhou Y, Hu X, Shao C. MicroRNA 483-3p suppresses the expression of DPC4/Smad4 in pancreatic cancer. FEBS Lett. 2011;585:207–13.
Xia X, Zhang K, Cen G, Jiang T, Cao J, Huang K, et al. MicroRNA-301a-3p promotes pancreatic cancer progression via negative regulation of SMAD4. Oncotarget. 2015;6:21046–63.
Li L, Li Z, Kong X, Xie D, Jia Z, Jiang W, et al. Down-regulation of microRNA-494 via loss of SMAD4 increases FOXM1 and β-catenin signaling in pancreatic ductal adenocarcinoma cells. Gastroenterology. 2014;147:485–97.
Zhong X, Chung AC, Chen HY, Meng XM, Lan HY. Smad3-mediated upregulation of miR-21 promotes renal fibrosis. J Am Soc Nephrol. 2011;22:1668–81.
Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA- mediated microRNA maturation. Nature. 2008;454:56–61.
Sempere LF, Preis M, Yezefski T, Ouyang H, Suriawinata AA, Silahtaroglu A, et al. Fluorescence-based codetection with protein markers reveals distinct cellular compartments for altered MicroRNA expression in solid tumors. Clin Cancer Res. 2010;16:4246–55.
Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90.
Thornley JA, Trask HW, Ringelberg CS, Ridley CJ, Wang S, Sal-Lari RC, et al. SMAD4-dependent polysome RNA recruitment in human pancreatic cancer cells. Mol Carcinog. 2012;51:771–82.
Wang J, Shao N, Ding X, Tan B, Song Q, Wang N, et al. Crosstalk between transforming growth factor-β signaling pathway and long non-coding RNAs in cancer. Cancer Lett. 2016;370:296–301.
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–81.
Xu X, Brodie SG, Yang X, Im YH, Parks WT, Chen L, et al. Haploid loss of the tumor suppressor Smad4/Dpc4 initiates gastric polyposis and cancer in mice. Oncogene. 2000;19:1868–74.
Sirard C, de la Pompa JL, Elia A, Itie A, Mirtsos C, Cheung A, et al. The tumor suppressor gene Smad4/Dpc4 is required for gastrulation and later for anterior development of the mouse embryo. Genes Dev. 1998;12:107–19.
Duda DG, Sunamura M, Lefter LP, Furukawa T, Yokoyama T, Yatsuoka T, et al. Restoration of SMAD4 by gene therapy reverses the invasive phenotype in pancreatic adenocarcinoma cells. Oncogene. 2003;22:6857–64.
Schwarte-Waldhoff I, Volpert OV, Bouck NP, Sipos B, Hahn SA, Klein-Scory S, et al. Smad4/DPC4-mediated tumor suppression through suppression of angiogenesis. Proc Natl Acad Sci USA. 2000;97:9624–9.
Yasutome M, Gunn J, Korc M. Restoration of Smad4 in BxPC3 pancreatic cancer cells attenuates proliferation without altering angiogenesis. Clin Exp Metastasis. 2005;22:461–73.
Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–50.
Pérez-Mancera PA, Guerra C, Barbacid M, Tuveson DA. What we have learned about pancreatic cancer from mouse models. Gastroenterology. 2012;142:1079–92.
Bardeesy N, Cheng KH, Berger JH, Chu GC, Pahler J, Olson P, et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20:3130–46.
Ardito CM, Grüner BM, Takeuchi KK, Lubeseder-Martellato C, Teichmann N, Mazur PK, et al. EGF receptor is required for KRAS-induced pancreatic tumorigenesis. Cancer Cell. 2012;22:304–17.
Norris AM, Gore A, Balboni A, Young A, Longnecker DS, Korc M. AGR2 is a SMAD4-suppressible gene that modulates MUC1 levels and promotes the initiation and progression of pancreatic intraepithelial neoplasia. Oncogene. 2013;32:3867–76.
Lin L, Tu HB, Wu L, Liu M, Jiang GN. MicroRNA-21 regulates non-small cell lung cancer cell invasion and chemo-sensitivity through SMAD7. Cell Physiol Biochem. 2016;38:2152–62.
Smith AL, Iwanaga R, Drasin DJ, Micalizzi DS, Vartuli RL, Tan AC, et al. The miR-106b-25 cluster targets Smad7, activates TGF-β signaling, and induces EMT and tumor initiating cell characteristics downstream of Six1 in human breast cancer. Oncogene. 2012;31:5162–71.
Yu J, Lei R, Zhuang X, Li X, Li G, Lev S, et al. MicroRNA-182 targets SMAD7 to potentiate TGFβ-induced epithelial-mesenchymal transition and metastasis of cancer cells. Nat Commun. 2016;7:13884. doi:10.1038/ncomms13884.
Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, Derynck R, et al. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biothechnol. 2011;29:443–8.
Yan X, Chen YG. Smad7: not only a regulator, but also a cross-talk mediator of TGF-β signalling. Biochem J. 2011;434:1–10.
Arnold NB, Ketterer K, Kleeff J, Friess H, Buchler MW, Korc M. Thioredoxin is downstream of Smad7 in a pathway that promotes growth and suppresses cisplatin-induced apoptosis in pancreatic cancer. Cancer Res. 2004;64:3599–606.
Boyer Arnold N, Korc M. Smad7 abrogates transforming growth factor-beta1-mediated growth inhibition in COLO-357 cells through functional inactivation of the retinoblastoma protein. J Biol Chem. 2005;280:21858–66.
Friess H, Yamanaka Y, Buchler M, Ebert M, Beger HG, Gold LI, et al. Enhanced expression of transforming growth factor beta isoforms in pancreatic cancer correlates with decreased survival. Gastroenterology. 1993;105:1846–56.
Aikawa T, Gunn J, Spong SM, Klaus SJ, Korc M. Connective tissue growth factor-specific antibody attenuates tumor growth, metastasis, and angiogenesis in an orthotopic mouse model of pancreatic cancer. Mol Cancer Ther. 2006;5:1108–16.
Neesse A, Frese KK, Bapiro TE, Nakagawa T, Sternlicht MD, Seeley TW, et al. CTGF antagonism with mAb FG-3019 enhances chemotherapy response without increasing drug delivery in murine ductal pancreas cancer. Proc Natl Acad Sci USA. 2013;110:12325–30.
Lawler S, Feng XH, Chen RH, Maruoka EM, Turck CW, Griswold-Prenner I, et al. The type II transforming growth factor-beta receptor autophosphorylates not only on serine and threonine but also on tyrosine residues. J Biol Chem. 1997;272:14850–9.
Lee MK, Pardoux C, Hall MC, Lee PS, Warburton D, Qing J, et al. TGF-beta activates Erk MAP kinase signalling through direct phosphorylation of ShcA. EMBO J. 2007;26:3957–67.
Sorrentino A, Thakur N, Grimsby S, Marcusson A, von Bulow V, Schuster N, et al. The type I TGF-beta receptor engages TRAF6 to activate TAK1 in a receptor kinase-independent manner. Nat Cell Biol. 2008;10:1199–207.
Zhong Y, Naito Y, Cope L, Naranjo-Suarez S, Saunders T, Hong SM, et al. Functional p38 MAPK identified by biomarker profiling of pancreatic cancer restrains growth through JNK inhibition and correlates with improved survival. Clin Cancer Res. 2014;20:6200–11.
Zhang H, Davies KJ, Forman HJ. TGFβ1 rapidly activates Src through a non-canonical redox signaling mechanism. Arch Biochem Biophys. 2015;568:1–7.
Hoefer M, Anderer FA. Anti-transforming growth factor beta antibodies with predefined specificity inhibit metastasis of highly tumorigenic human xenotransplants in nu/nu mice. Cancer Immunol Immunother. 1995;41:302–8.
Marzo AL, Fitzpatrick DR, Robinson BW, Scott B. Antisense oligonucleotides specific for transforming growth factor beta2 inhibit the growth of malignant mesothelioma both in vitro and in vivo. Cancer Res. 1997;57:3200–7.
Lopez AR, Cook J, Deininger PL, Derynck R. Dominant negative mutants of transforming growth factor-beta 1 inhibit the secretion of different transforming growth factor-beta isoforms. Mol Cell Biol. 1992;12:1674–9.
Rowland-Goldsmith MA, Maruyama H, Kusama T, Ralli S, Korc M. Soluble type II transforming growth factor-beta (TGF-beta) receptor inhibits TGF-beta signaling in COLO-357 pancreatic cancer cells in vitro and attenuates tumor formation. Clin Cancer Res. 2001;7:2931–40.
Nam J-S, Terabe M, Mamura M, Kang M-J, Chae H, Stuelten C, et al. An anti-transforming growth factor β antibody suppresses metastasis via cooperative effects on multiple cell compartments. Cancer Res. 2008;68:3835–43.
Connolly EC, Saunier EF, Quigley D, Luu MT, De Sapio A, Hann B, et al. Outgrowth of drug-resistant carcinomas expressing markers of tumor aggression after long-term TβRI/II kinase inhibition with LY2109761. Cancer Res. 2011;71:2339–49.
Hezel AF, Deshpande V, Zimmerman SM, Contino G, Alagesan B, O’Dell MR, et al. TGF-β and αvβ6 integrin act in a common pathway to suppress pancreatic cancer progression. Cancer Res. 2012;72:4840–5.
David CJ, Huang YH, Chen M, Su J, Zou Y, Bardeesy N, et al. TGF-β tumor suppression through a lethal EMT. Cell. 2016;164:1015–30.
Craven KE, Gore J, Wilson JL, Korc M. Angiogenic gene signature in human pancreatic cancer correlates with TGF-beta and inflammatory transcriptomes. Oncotarget. 2016;7:323–41.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Section Editor information
Rights and permissions
Copyright information
© 2017 Springer Science+Business Media LLC
About this entry
Cite this entry
Korc, M. (2017). Smad4-TGF-β Signaling Pathways in Pancreatic Cancer Pathogenesis. In: Neoptolemos, J., Urrutia, R., Abbruzzese, J., Büchler, M. (eds) Pancreatic Cancer. Springer, New York, NY. https://doi.org/10.1007/978-1-4939-6631-8_17-2
Download citation
DOI: https://doi.org/10.1007/978-1-4939-6631-8_17-2
Received:
Accepted:
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4939-6631-8
Online ISBN: 978-1-4939-6631-8
eBook Packages: Springer Reference Biomedicine and Life SciencesReference Module Biomedical and Life Sciences