Abstract
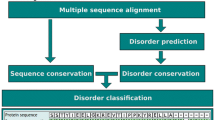
Studying the evolutionary conservation of proteins can be a valuable tool for understanding its function. At the sequence level, the conservation of each residue can be used to infer the importance of the particular regions of proteins. In the case of protein disulphide bonds, the conservation of the cysteines involved can be used to infer the conservation of the disulphide bond itself. In this chapter, bioinformatics methods are described that can be used to assess the conservation of a protein disulphide bond with a focus on the study of human proteins. Conservation will be assessed at the species and at the human population level. The methods described make use of publicly available databases and can be applied by any researcher using a standard desktop computer with Internet access.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Poole LB (2015) The basics of thiols and cysteines in redox biology and chemistry. Free Radic Biol Med 80:148–157. https://doi.org/10.1016/j.freeradbiomed.2014.11.013
Fass D (2012) Disulfide bonding in protein biophysics. Annu Rev Biophys 41:63–79. https://doi.org/10.1146/annurev-biophys-050511-102321
Butera D, Cook KM, Chiu J, Wong JW, Hogg PJ (2014) Control of blood proteins by functional disulfide bonds. Blood 123(13):2000–2007. https://doi.org/10.1182/blood-2014-01-549816
Wong JW, Ho SY, Hogg PJ (2011) Disulfide bond acquisition through eukaryotic protein evolution. Mol Biol Evol 28(1):327–334. https://doi.org/10.1093/molbev/msq194
Matthias LJ, Yam PT, Jiang XM, Vandegraaff N, Li P, Poumbourios P, Donoghue N, Hogg PJ (2002) Disulfide exchange in domain 2 of CD4 is required for entry of HIV-1. Nat Immunol 3(8):727–732. https://doi.org/10.1038/ni815
International HapMap C (2005) A haplotype map of the human genome. Nature 437(7063):1299–1320. https://doi.org/10.1038/nature04226
UniProt Consortium T (2018) UniProt: the universal protein knowledgebase. Nucleic Acids Res 46(5):2699. https://doi.org/10.1093/nar/gky092
Altenhoff AM, Glover NM, Train CM, Kaleb K, Warwick Vesztrocy A, Dylus D, de Farias TM, Zile K, Stevenson C, Long J, Redestig H, Gonnet GH, Dessimoz C (2018) The OMA orthology database in 2018: retrieving evolutionary relationships among all domains of life through richer web and programmatic interfaces. Nucleic Acids Res 46(D1):D477–D485. https://doi.org/10.1093/nar/gkx1019
Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ (2009) Jalview version 2 – a multiple sequence alignment editor and analysis workbench. Bioinformatics 25(9):1189–1191. https://doi.org/10.1093/bioinformatics/btp033
Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won HH, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG, Exome Aggregation Consortium (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature 536(7616):285–291. https://doi.org/10.1038/nature19057
Rosenbloom KR, Armstrong J, Barber GP, Casper J, Clawson H, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, Harte RA, Heitner S, Hickey G, Hinrichs AS, Hubley R, Karolchik D, Learned K, Lee BT, Li CH, Miga KH, Nguyen N, Paten B, Raney BJ, Smit AF, Speir ML, Zweig AS, Haussler D, Kuhn RM, Kent WJ (2015) The UCSC genome browser database: 2015 update. Nucleic Acids Res 43(Database issue):D670–D681. https://doi.org/10.1093/nar/gku1177
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5):1792–1797. https://doi.org/10.1093/nar/gkh340
Thompson JD, Linard B, Lecompte O, Poch O (2011) A comprehensive benchmark study of multiple sequence alignment methods: current challenges and future perspectives. PLoS One 6(3):e18093. https://doi.org/10.1371/journal.pone.0018093
Letunic I, Bork P (2016) Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44(W1):W242–W245. https://doi.org/10.1093/nar/gkw290
Acknowledgments
This work is supported by an Australian Research Council Future Fellowship (FT130100096) to JWHW.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Wong, J.W.H. (2019). Assessing the Evolutionary Conservation of Protein Disulphide Bonds. In: Hogg, P. (eds) Functional Disulphide Bonds. Methods in Molecular Biology, vol 1967. Humana, New York, NY. https://doi.org/10.1007/978-1-4939-9187-7_2
Download citation
DOI: https://doi.org/10.1007/978-1-4939-9187-7_2
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-4939-9186-0
Online ISBN: 978-1-4939-9187-7
eBook Packages: Springer Protocols