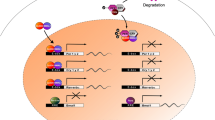
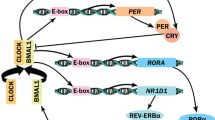
Abstract
Self-sustained and synchronized to environmental stimuli, circadian clocks are under genetic and epigenetic regulation. Recent findings have greatly increased our understanding of epigenetic plasticity governed by circadian clock. Thus, the link between circadian clock and epigenetic machinery is reciprocal. Circadian clock can affect epigenetic features including genomic DNA methylation, noncoding RNA, mainly miRNA expression, and histone modifications resulted in their 24-h rhythms. Concomitantly, these epigenetic events can directly modulate cyclic system of transcription and translation of core circadian genes and indirectly clock output genes. Significant findings interlocking circadian clock, epigenetics, and cancer have been revealed, particularly in breast, colorectal, and blood cancers. Aberrant methylation of circadian gene promoter regions and miRNA expression affected circadian gene expression, together with 24-h expression oscillation pace have been frequently observed.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Change history
09 March 2019
The Chapter was inadvertently published with incorrect figure (Fig. 1). The same has been corrected in the chapter.
References
Terzibasi-Tozzini E, Martinez-Nicolas A, Lucas-Sanchez A (2017) The clock is ticking. Ageing of the circadian system: from physiology to cell cycle. Semin Cell Dev Biol 70:164–176. https://doi.org/10.1016/j.semcdb.2017.06.011
Webb AB, Oates AC (2016) Timing by rhythms: daily clocks and developmental rulers. Dev Growth Differ 58(1):43–58. https://doi.org/10.1111/dgd.12242
Delezie J, Challet E (2011) Interactions between metabolism and circadian clocks: reciprocal disturbances. Ann N Y Acad Sci 1243:30–46
Reppert SM, Weaver DR (2001) Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol 63:647–676
Ukai H, Ueda HR (2010) Systems biology of mammalian circadian clocks. Annu Rev Physiol 72:579–603
Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ (1998) Role of the CLOCK protein in the mammalian circadian mechanism. Science 280(5369):1564–1569
Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA (2000) Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103(7):1009–1017
Jin X, Shearman LP, Weaver DR, Zylka MJ, de Vries GJ, Reppert SM (1999) A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell 96(1):57–68
van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D, Buijs R, Bootsma D, Hoeijmakers JH, Yasui A (1999) Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398(6728):627–630
Hogenesch JB, Gu YZ, Moran SM, Shimomura K, Radcliffe LA, Takahashi JS, Bradfield CA (2000) The basic helix-loop-helix-PAS protein MOP9 is a brain-specific heterodimeric partner of circadian and hypoxia factors. J Neurosci 20(13):Rc83
Zhou YD, Barnard M, Tian H, Li X, Ring HZ, Francke U, Shelton J, Richardson J, Russell DW, McKnight SL (1997) Molecular characterization of two mammalian bHLH-PAS domain proteins selectively expressed in the central nervous system. Proc Natl Acad Sci U S A 94(2):713–718
Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U (2002) The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110(2):251–260
Akashi M, Takumi T (2005) The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat Struct Mol Biol 12(5):441–448
Stratmann M, Schibler U (2006) Properties, entrainment, and physiological functions of mammalian peripheral oscillators. J Biol Rhythm 21(6):494–506
Ukai-Tadenuma M, Yamada RG, Xu H, Ripperger JA, Liu AC, Ueda HR (2011) Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell 144(2):268–281
Korencic A, Bordyugov G, Kosir R, Rozman D, Golicnik M, Herzel H (2012) The interplay of cis-regulatory elements rules circadian rhythms in mouse liver. PLoS One 7(11):e46835
Korencic A, Kosir R, Bordyugov G, Lehmann R, Rozman D, Herzel H (2014) Timing of circadian genes in mammalian tissues. Sci Rep 4:5782. https://doi.org/10.1038/srep05782
Pett JP, Korencic A, Wesener F, Kramer A, Herzel H (2016) Feedback loops of the mammalian circadian clock constitute Repressilator. PLoS Comput Biol 12(12):e1005266. https://doi.org/10.1371/journal.pcbi.1005266
Akashi M, Okamoto A, Tsuchiya Y, Todo T, Nishida E, Node K (2014) A positive role for PERIOD in mammalian circadian gene expression. Cell Rep 7(4):1056–1064
Mitsui S, Yamaguchi S, Matsuo T, Ishida Y, Okamura H (2001) Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Genes Dev 15(8):995–1006
Yamaguchi S, Mitsui S, Yan L, Yagita K, Miyake S, Okamura H (2000) Role of DBP in the circadian oscillatory mechanism. Mol Cell Biol 20(13):4773–4781
Ko CH, Takahashi JS (2006) Molecular components of the mammalian circadian clock. Hum Mol Genet 15:R271–R277 Spec No 2. https://doi.org/10.1093/hmg/ddl207
Stojkovic K, Wing SS, Cermakian N (2014) A central role for ubiquitination within a circadian clock protein modification code. Front Mol Neurosci 7:69. https://doi.org/10.3389/fnmol.2014.00069
Mazzoccoli G, Laukkanen MO, Vinciguerra M, Colangelo T, Colantuoni V (2016) A timeless link between circadian patterns and disease. Trends Mol Med 22(1):68–81
Hatanaka F, Matsubara C, Myung J, Yoritaka T, Kamimura N, Tsutsumi S, Kanai A, Suzuki Y, Sassone-Corsi P, Aburatani H, Sugano S, Takumi T (2010) Genome-wide profiling of the core clock protein BMAL1 targets reveals a strict relationship with metabolism. Mol Cell Biol 30(24):5636–5648
Yi CH, Zheng T, Leaderer D, Hoffman A, Zhu Y (2009) Cancer-related transcriptional targets of the circadian gene NPAS2 identified by genome-wide ChIP-on-chip analysis. Cancer Lett 284(2):149–156
Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, Naef F (2011) Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol 9(2):e1000595. https://doi.org/10.1371/journal.pbio.1000595
Menet JS, Pescatore S, Rosbash M (2014) CLOCK:BMAL1 is a pioneer-like transcription factor. Genes Dev 28(1. United States):8–13. https://doi.org/10.1101/gad.228536.113
Asher G, Schibler U (2011) Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab 13(2):125–137
Eckel-Mahan K, Sassone-Corsi P (2013) Metabolism and the circadian clock converge. Physiol Rev 93(1):107–135
Ptitsyn AA, Zvonic S, Conrad SA, Scott LK, Mynatt RL, Gimble JM (2006) Circadian clocks are resounding in peripheral tissues. PLoS Comput Biol 2(3):e16. https://doi.org/10.1371/journal.pcbi.0020016
Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB (2002) Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109(3):307–320
Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, Kyriacou CP (2002) Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol 12(7):540–550
Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB (2014) A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci U S A 111(45):16219–16224
Archer SN, Laing EE, Moller-Levet CS, van der Veen DR, Bucca G, Lazar AS, Santhi N, Slak A, Kabiljo R, von Schantz M, Smith CP, Dijk DJ (2014) Mistimed sleep disrupts circadian regulation of the human transcriptome. Proc Natl Acad Sci U S A 111(6):E682–E691
Moller-Levet CS, Archer SN, Bucca G, Laing EE, Slak A, Kabiljo R, Lo JC, Santhi N, von Schantz M, Smith CP, Dijk DJ (2013) Effects of insufficient sleep on circadian rhythmicity and expression amplitude of the human blood transcriptome. Proc Natl Acad Sci U S A 110(12):E1132–E1141
Simo-Riudalbas L, Esteller M (2014) Cancer genomics identifies disrupted epigenetic genes. Hum Genet 133(6):713–725
Powell WT, LaSalle JM (2015) Epigenetic mechanisms in diurnal cycles of metabolism and neurodevelopment. Hum Mol Genet 24(R1):R1–R9. https://doi.org/10.1093/hmg/ddv234
Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS (2012) Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338(6105):349–354
Brown SE, Fraga MF, Weaver IC, Berdasco M, Szyf M (2007) Variations in DNA methylation patterns during the cell cycle of HeLa cells. Epigenetics 2(1):54–65
Bonsch D, Hothorn T, Krieglstein C, Koch M, Nehmer C, Lenz B, Reulbach U, Kornhuber J, Bleich S (2007) Daily variations of homocysteine concentration may influence methylation of DNA in normal healthy individuals. Chronobiol Int 24(2):315–326
Xia L, Ma S, Zhang Y, Wang T, Zhou M, Wang Z, Zhang J (2015) Daily variation in global and local DNA methylation in mouse livers. PLoS One 10(2):e0118101. https://doi.org/10.1371/journal.pone.0118101
Azzi A, Dallmann R, Casserly A, Rehrauer H, Patrignani A, Maier B, Kramer A, Brown SA (2014) Circadian behavior is light-reprogrammed by plastic DNA methylation. Nat Neurosci 17(3):377–382
Vollmers C, Schmitz RJ, Nathanson J, Yeo G, Ecker JR, Panda S (2012) Circadian oscillations of protein-coding and regulatory RNAs in a highly dynamic mammalian liver epigenome. Cell Metab 16(6):833–845
Massart R, Freyburger M, Suderman M, Paquet J, El Helou J, Belanger-Nelson E, Rachalski A, Koumar OC, Carrier J, Szyf M, Mongrain V (2014) The genome-wide landscape of DNA methylation and hydroxymethylation in response to sleep deprivation impacts on synaptic plasticity genes. Transl Psychiatry 4:e347. https://doi.org/10.1038/tp.2013.120
Zhu Y, Stevens RG, Hoffman AE, Tjonneland A, Vogel UB, Zheng T, Hansen J (2011) Epigenetic impact of long-term shiftwork: pilot evidence from circadian genes and whole-genome methylation analysis. Chronobiol Int 28(10):852–861
Bhatti P, Zhang Y, Song X, Makar KW, Sather CL, Kelsey KT, Houseman EA, Wang P (2014) Nightshift work and genome-wide DNA methylation. Chronobiol Int 32:103–112. https://doi.org/10.3109/07420528.2014.956362
Schwimmer H, Metzer A, Pilosof Y, Szyf M, Machnes ZM, Fares F, Harel O, Haim A (2014) Light at night and melatonin have opposite effects on breast cancer tumors in mice assessed by growth rates and global DNA methylation. Chronobiol Int 31(1):144–150
Lim AS, Srivastava GP, Yu L, Chibnik LB, Xu J, Buchman AS, Schneider JA, Myers AJ, Bennett DA, De Jager PL (2014) 24-hour rhythms of DNA methylation and their relation with rhythms of RNA expression in the human dorsolateral prefrontal cortex. PLoS Genet 10(11):e1004792. https://doi.org/10.1371/journal.pgen.1004792
Krishnaiah SY, Wu G, Altman BJ, Growe J, Rhoades SD, Coldren F, Venkataraman A, Olarerin-George AO, Francey LJ, Mukherjee S, Girish S, Selby CP, Cal S, Er U, Sianati B, Sengupta A, Anafi RC, Kavakli IH, Sancar A, Baur JA, Dang CV, Hogenesch JB, Weljie AM (2017) Clock regulation of metabolites reveals coupling between transcription and metabolism. Cell Metab 25(4):961–974.e964
Flavahan WA, Gaskell E, Bernstein BE (2017) Epigenetic plasticity and the hallmarks of cancer. Science 357(6348):eaal2380. https://doi.org/10.1126/science.aal2380
Unsal-Kaçmaz K, Mullen TE, Kaufmann WK, Sancar A (2005) Coupling of human circadian and cell cycles by the timeless protein. Mol Cell Biol 25(8):3109–3116
Gauger MA, Sancar A (2005) Cryptochrome, circadian cycle, cell cycle checkpoints, and cancer. Cancer Res 65(15):6828–6834
Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H (2003) Control mechanism of the circadian clock for timing of cell division in vivo. Science 302(5643):255–259
Kettner NM, Katchy CA, Fu L (2014) Circadian gene variants in cancer. Ann Med 46(4):208–220
Mao Y, Fu A, Hoffman AE, Jacobs DI, Jin M, Chen K, Zhu Y (2015) The circadian gene CRY2 is associated with breast cancer aggressiveness possibly via epigenomic modifications. Tumour Biol 36(5):3533–3539
Chen ST, Choo KB, Hou MF, Yeh KT, Kuo SJ, Chang JG (2005) Deregulated expression of the PER1, PER2 and PER3 genes in breast cancers. Carcinogenesis 26(7):1241–1246
Kuo SJ, Chen ST, Yeh KT, Hou MF, Chang YS, Hsu NC, Chang JG (2009) Disturbance of circadian gene expression in breast cancer. Virchows Arch 454(4):467–474
Hoffman AE, Zheng T, Yi CH, Stevens RG, Ba Y, Zhang Y, Leaderer D, Holford T, Hansen J, Zhu Y (2010) The core circadian gene Cryptochrome 2 influences breast cancer risk, possibly by mediating hormone signaling. Cancer Prev Res (Phila) 3(4):539–548
Li L, Lee KM, Han W, Choi JY, Lee JY, Kang GH, Park SK, Noh DY, Yoo KY, Kang D (2010) Estrogen and progesterone receptor status affect genome-wide DNA methylation profile in breast cancer. Hum Mol Genet 19(21):4273–4277
Liu L, Shen H, Wang Y (2017) CRY2 is suppressed by FOXM1 mediated promoter hypermethylation in breast cancer. Biochem Biophys Res Commun 490(1):44–50
Hoffman AE, Yi CH, Zheng T, Stevens RG, Leaderer D, Zhang Y, Holford TR, Hansen J, Paulson J, Zhu Y (2010) CLOCK in breast tumorigenesis: genetic, epigenetic, and transcriptional profiling analyses. Cancer Res 70(4):1459–1468
Hoffman AE, Zheng T, Ba Y, Stevens RG, Yi CH, Leaderer D, Zhu Y (2010) Phenotypic effects of the circadian gene Cryptochrome 2 on cancer-related pathways. BMC Cancer 10:110. https://doi.org/10.1186/1471-2407-10-110
Fu A, Leaderer D, Zheng T, Hoffman AE, Stevens RG, Zhu Y (2012) Genetic and epigenetic associations of circadian gene TIMELESS and breast cancer risk. Mol Carcinog 51(12):923–929
Yang M, Kim HS, Cho MY (2014) Different methylation profiles between intestinal and diffuse sporadic gastric carcinogenesis. Clin Res Hepatol Gastroenterol 38(5):613–620
Tahara T, Maegawa S, Chung W, Garriga J, Jelinek J, Estecio MR, Shibata T, Hirata I, Arisawa T, Issa JP (2013) Examination of whole blood DNA methylation as a potential risk marker for gastric cancer. Cancer Prev Res (Phila) 6(10):1093–1100
Alexander M, Burch JB, Steck SE, Chen CF, Hurley TG, Cavicchia P, Shivappa N, Guess J, Zhang H, Youngstedt SD, Creek KE, Lloyd S, Jones K, Hebert JR (2017) Case-control study of candidate gene methylation and adenomatous polyp formation. Int J Color Dis 32(2):183–192
Tomita T, Kurita R, Onishi Y (2017) Epigenetic regulation of the circadian clock: role of 5-aza-2′-deoxycytidine. Biosci Rep 37(3):BSR20170053. https://doi.org/10.1042/bsr20170053
Hsu MC, Huang CC, Choo KB, Huang CJ (2007) Uncoupling of promoter methylation and expression of Period1 in cervical cancer cells. Biochem Biophys Res Commun 360(1):257–262
Shih MC, Yeh KT, Tang KP, Chen JC, Chang JG (2006) Promoter methylation in circadian genes of endometrial cancers detected by methylation-specific PCR. Mol Carcinog 45(10):732–740
Yeh KT, Yang MY, Liu TC, Chen JC, Chan WL, Lin SF, Chang JG (2005) Abnormal expression of period 1 (PER1) in endometrial carcinoma. J Pathol 206(1):111–120
Neumann O, Kesselmeier M, Geffers R, Pellegrino R, Radlwimmer B, Hoffmann K, Ehemann V, Schemmer P, Schirmacher P, Lorenzo Bermejo J, Longerich T (2012) Methylome analysis and integrative profiling of human HCCs identify novel protumorigenic factors. Hepatology 56(5):1817–1827
Lin YM, Chang JH, Yeh KT, Yang MY, Liu TC, Lin SF, Su WW, Chang JG (2008) Disturbance of circadian gene expression in hepatocellular carcinoma. Mol Carcinog 47(12):925–933
Gery S, Komatsu N, Kawamata N, Miller CW, Desmond J, Virk RK, Marchevsky A, McKenna R, Taguchi H, Koeffler HP (2007) Epigenetic silencing of the candidate tumor suppressor gene Per1 in non-small cell lung cancer. Clin Cancer Res 13(5):1399–1404
Wang F, Luo Y, Li C, Chen L (2014) Correlation between deregulated expression of PER2 gene and degree of glioma malignancy. Tumori 100(6):e266–e272. https://doi.org/10.1700/1778.19292
Fan W, Chen X, Li C, Chen L, Liu P, Chen Z (2014) The analysis of deregulated expression and methylation of the PER2 genes in gliomas. J Cancer Res Ther 10(3):636–640
Hanoun M, Eisele L, Suzuki M, Greally JM, Huttmann A, Aydin S, Scholtysik R, Klein-Hitpass L, Duhrsen U, Durig J (2012) Epigenetic silencing of the circadian clock gene CRY1 is associated with an indolent clinical course in chronic lymphocytic leukemia. PLoS One 7(3):e34347. https://doi.org/10.1371/journal.pone.0034347
Taniguchi H, Fernandez AF, Setien F, Ropero S, Ballestar E, Villanueva A, Yamamoto H, Imai K, Shinomura Y, Esteller M (2009) Epigenetic inactivation of the circadian clock gene BMAL1 in hematologic malignancies. Cancer Res 69(21):8447–8454
Yang MY, Chang JG, Lin PM, Tang KP, Chen YH, Lin HY, Liu TC, Hsiao HH, Liu YC, Lin SF (2006) Downregulation of circadian clock genes in chronic myeloid leukemia: alternative methylation pattern of hPER3. Cancer Sci 97(12):1298–1307
Yeh CM, Shay J, Zeng TC, Chou JL, Huang TH, Lai HC, Chan MW (2014) Epigenetic silencing of ARNTL, a circadian gene and potential tumor suppressor in ovarian cancer. Int J Oncol 45(5):2101–2107
Nakatome M, Orii M, Hamajima M, Hirata Y, Uemura M, Hirayama S, Isobe I (2011) Methylation analysis of circadian clock gene promoters in forensic autopsy specimens. Legal medicine (Tokyo, Japan) 13(4):205–209
Biswas S, Rao CM (2017) Epigenetics in cancer: fundamentals and beyond. Pharmacol Ther. https://doi.org/10.1016/j.pharmthera.2017.02.011
Liz J, Esteller M (2016) lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta 1859(1):169–176
Na YJ, Sung JH, Lee SC, Lee YJ, Choi YJ, Park WY, Shin HS, Kim JH (2009) Comprehensive analysis of microRNA-mRNA co-expression in circadian rhythm. Exp Mol Med 41(9):638–647
Cheng HY, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, Obrietan K (2007) microRNA modulation of circadian-clock period and entrainment. Neuron 54(5):813–829
Alvarez-Saavedra M, Antoun G, Yanagiya A, Oliva-Hernandez R, Cornejo-Palma D, Perez-Iratxeta C, Sonenberg N, Cheng HY (2011) miRNA-132 orchestrates chromatin remodeling and translational control of the circadian clock. Hum Mol Genet 20(4):731–751
Nagel R, Clijsters L, Agami R (2009) The miRNA-192/194 cluster regulates the period gene family and the circadian clock. FEBS J 276(19):5447–5455
Chen R, D'Alessandro M, Lee C (2013) miRNAs are required for generating a time delay critical for the circadian oscillator. Curr Biol 23(20):1959–1968
Zhou W, Li Y, Wang X, Wu L, Wang Y (2011) MiR-206-mediated dynamic mechanism of the mammalian circadian clock. BMC Syst Biol 5:141. https://doi.org/10.1186/1752-0509-5-141
Chacolla-Huaringa R, Moreno-Cuevas J, Trevino V, Scott SP (2017) Entrainment of breast cell lines results in rhythmic fluctuations of MicroRNAs. Int J Mol Sci 18:7. https://doi.org/10.3390/ijms18071499
Kochan DZ, Ilnytskyy Y, Golubov A, Deibel SH, McDonald RJ, Kovalchuk O (2015) Circadian disruption-induced microRNAome deregulation in rat mammary gland tissues. Oncoscience 2(4):428–442
Han Y, Meng F, Venter J, Wu N, Wan Y, Standeford H, Francis H, Meininger C, Greene J Jr, Trzeciakowski JP, Ehrlich L, Glaser S, Alpini G (2016) miR-34a-dependent overexpression of Per1 decreases cholangiocarcinoma growth. J Hepatol 64(6):1295–1304
Shi F, Chen X, Fu A, Hansen J, Stevens R, Tjonneland A, Vogel UB, Zheng T, Zhu Y (2013) Aberrant DNA methylation of miR-219 promoter in long-term night shiftworkers. Environ Mol Mutagen 54(6):406–413
Lee SE, Kim SJ, Youn JP, Hwang SY, Park CS, Park YS (2011) MicroRNA and gene expression analysis of melatonin-exposed human breast cancer cell lines indicating involvement of the anticancer effect. J Pineal Res 51(3):345–352
Mazzoccoli G, Colangelo T, Panza A, Rubino R, Tiberio C, Palumbo O, Carella M, Trombetta D, Gentile A, Tavano F, Valvano MR, Storlazzi CT, Macchia G, De Cata A, Bisceglia G, Capocefalo D, Colantuoni V, Sabatino L, Piepoli A, Mazza T (2016) Analysis of clock gene-miRNA correlation networks reveals candidate drivers in colorectal cancer. Oncotarget 7(29):45444–45461
Li A, Lin X, Tan X, Yin B, Han W, Zhao J, Yuan J, Qiang B, Peng X (2013) Circadian gene clock contributes to cell proliferation and migration of glioma and is directly regulated by tumor-suppressive miR-124. FEBS Lett 587(15):2455–2460
Wu S, Fesler A, Ju J (2016) Implications of circadian rhythm regulation by microRNAs in colorectal Cancer. Cancer Transl Med 2(1):1–6
Hong Z, Feng Z, Sai Z, Tao S (2014) PER3, a novel target of miR-103, plays a suppressive role in colorectal cancer in vitro. BMB Rep 47(9):500–505
Cui M, Zheng M, Sun B, Wang Y, Ye L, Zhang X (2015) A long noncoding RNA perturbs the circadian rhythm of hepatoma cells to facilitate hepatocarcinogenesis. Neoplasia 17(1):79–88
Etchegaray JP, Lee C, Wade PA, Reppert SM (2003) Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature 421(6919):177–182
Valekunja UK, Edgar RS, Oklejewicz M, van der Horst GT, O'Neill JS, Tamanini F, Turner DJ, Reddy AB (2013) Histone methyltransferase MLL3 contributes to genome-scale circadian transcription. Proc Natl Acad Sci U S A 110(4):1554–1559
Crosio C, Cermakian N, Allis CD, Sassone-Corsi P (2000) Light induces chromatin modification in cells of the mammalian circadian clock. Nat Neurosci 3(12):1241–1247
Naruse Y, Oh-hashi K, Iijima N, Naruse M, Yoshioka H, Tanaka M (2004) Circadian and light-induced transcription of clock gene Per1 depends on histone acetylation and deacetylation. Mol Cell Biol 24(14):6278–6287
Curtis AM, Seo SB, Westgate EJ, Rudic RD, Smyth EM, Chakravarti D, FitzGerald GA, McNamara P (2004) Histone acetyltransferase-dependent chromatin remodeling and the vascular clock. J Biol Chem 279(8):7091–7097
Katada S, Sassone-Corsi P (2010) The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat Struct Mol Biol 17(12):1414–1421
Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U (2008) SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134(2):317–328
Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P (2008) The NAD+−dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134(2):329–340
Doi M, Hirayama J, Sassone-Corsi P (2006) Circadian regulator CLOCK is a histone acetyltransferase. Cell 125(3):497–508
Hirayama J, Sahar S, Grimaldi B, Tamaru T, Takamatsu K, Nakahata Y, Sassone-Corsi P (2007) CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature 450:1086–1090
Rosenbloom KR, Armstrong J, Barber GP, Casper J, Clawson H, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, Harte RA, Heitner S, Hickey G, Hinrichs AS, Hubley R, Karolchik D, Learned K, Lee BT, Li CH, Miga KH, Nguyen N, Paten B, Raney BJ, Smit AF, Speir ML, Zweig AS, Haussler D, Kuhn RM, Kent WJ (2015) The UCSC genome browser database: 2015 update. Nucleic Acids Res 43(Database issue):D670–D681. https://doi.org/10.1093/nar/gku1177
Suzuki A, Wakaguri H, Yamashita R, Kawano S, Tsuchihara K, Sugano S, Suzuki Y, Nakai K (2015) DBTSS as an integrative platform for transcriptome, epigenome and genome sequence variation data. Nucleic Acids Res 43(Database issue):D87–D91. https://doi.org/10.1093/nar/gku1080
Yamashita R, Sugano S, Suzuki Y, Nakai K (2012) DBTSS: DataBase of transcriptional start sites progress report in 2012. Nucleic Acids Res 40(Database issue):D150–D154. https://doi.org/10.1093/nar/gkr1005
Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW (2013) GenBank. Nucleic Acids Res 41(Database issue):D36–D42. https://doi.org/10.1093/nar/gks1195
Dreos R, Ambrosini G, Cavin Perier R, Bucher P (2013) EPD and EPDnew, high-quality promoter resources in the next-generation sequencing era. Nucleic Acids Res 41(Database issue):D157–D164. https://doi.org/10.1093/nar/gks1233
Dreos R, Ambrosini G, Perier RC, Bucher P (2015) The eukaryotic promoter database: expansion of EPDnew and new promoter analysis tools. Nucleic Acids Res 43(Database issue):D92–D96. https://doi.org/10.1093/nar/gku1111
Abeel T, Van de Peer Y, Saeys Y (2009) Toward a gold standard for promoter prediction evaluation. Bioinformatics (Oxford, England) 25(12):i313–i320. https://doi.org/10.1093/bioinformatics/btp191
Li LC, Dahiya R (2002) MethPrimer: designing primers for methylation PCRs. Bioinformatics (Oxford, England) 18(11):1427–1431
McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, Cowley AP, Lopez R (2013) Analysis tool web services from the EMBL-EBI. Nucleic Acids Res 41(Web Server issue):W597–W600. https://doi.org/10.1093/nar/gkt376
Yan H, Tian S, Slager SL, Sun Z, Ordog T (2016) Genome-wide epigenetic studies in human disease: a primer on -Omic technologies. Am J Epidemiol 183(2):96–109
Knudsen S (1999) Promoter2.0: for the recognition of PolII promoter sequences. Bioinformatics (Oxford, England) 15(5):356–361
Halees AS, Leyfer D, Weng Z (2003) PromoSer: a large-scale mammalian promoter and transcription start site identification service. Nucleic Acids Res 31(13):3554–3559
Lee TY, Chang WC, Hsu JB, Chang TH, Shien DM (2012) GPMiner: an integrated system for mining combinatorial cis-regulatory elements in mammalian gene group. BMC Genomics 13(Suppl 1):S3. https://doi.org/10.1186/1471-2164-13-s1-s3
Zhang J, Shi Z, Nan Y, Li M (2016) Inhibiting malignant phenotypes of the bladder cancer cells by silencing long noncoding RNA SChLAP1. Int Urol Nephrol 48(5):711–716
Davuluri RV, Grosse I, Zhang MQ (2001) Computational identification of promoters and first exons in the human genome. Nat Genet 29(4):412–417
Down TA, Hubbard TJ (2002) Computational detection and location of transcription start sites in mammalian genomic DNA. Genome Res 12(3):458–461
Ponger L, Mouchiroud D (2002) CpGProD: identifying CpG islands associated with transcription start sites in large genomic mammalian sequences. Bioinformatics (Oxford, England) 18(4):631–633
Aranyi T, Tusnady GE (2007) BiSearch: ePCR tool for native or bisulfite-treated genomic template. Methods in molecular biology (Clifton, NJ) 402:385–402. https://doi.org/10.1007/978-1-59745-528-2_20
Schuffler P, Mikeska T, Waha A, Lengauer T, Bock C (2009) MethMarker: user-friendly design and optimization of gene-specific DNA methylation assays. Genome Biol 10(10):R105. https://doi.org/10.1186/gb-2009-10-10-r105
Pandey RV, Pulverer W, Kallmeyer R, Beikircher G, Pabinger S, Kriegner A, Weinhausel A (2016) MSP-HTPrimer: a high-throughput primer design tool to improve assay design for DNA methylation analysis in epigenetics. Clin Epigenetics 8:101. https://doi.org/10.1186/s13148-016-0269-3
Bock C, Reither S, Mikeska T, Paulsen M, Walter J, Lengauer T (2005) BiQ analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics (Oxford, England) 21(21):4067–4068
Lutsik P, Feuerbach L, Arand J, Lengauer T, Walter J, Bock C (2011) BiQ analyzer HT: locus-specific analysis of DNA methylation by high-throughput bisulfite sequencing. Nucleic Acids Res 39(Web Server issue):W551–W556. https://doi.org/10.1093/nar/gkr312
Becker D, Lutsik P, Ebert P, Bock C, Lengauer T, Walter J (2014) BiQ analyzer HiMod: an interactive software tool for high-throughput locus-specific analysis of 5-methylcytosine and its oxidized derivatives. Nucleic Acids Res 42(Web Server issue):W501–W507. https://doi.org/10.1093/nar/gku457
Xin Y, Chanrion B, O'Donnell AH, Milekic M, Costa R, Ge Y, Haghighi FG (2012) MethylomeDB: a database of DNA methylation profiles of the brain. Nucleic Acids Res 40(Database issue):D1245–D1249. https://doi.org/10.1093/nar/gkr1193
Lv J, Liu H, Su J, Wu X, Li B, Xiao X, Wang F, Wu Q, Zhang Y (2012) DiseaseMeth: a human disease methylation database. Nucleic Acids Res 40(Database issue):D1030–D1035. https://doi.org/10.1093/nar/gkr1169
Song Q, Decato B, Hong EE, Zhou M, Fang F, Qu J, Garvin T, Kessler M, Zhou J, Smith AD (2013) A reference methylome database and analysis pipeline to facilitate integrative and comparative epigenomics. PLoS One 8(12):e81148. https://doi.org/10.1371/journal.pone.0081148
Satterlee JS, Beckel-Mitchener A, McAllister K, Procaccini DC, Rutter JL, Tyson FL, Chadwick LH (2015) Community resources and technologies developed through the NIH roadmap Epigenomics program. Methods Mol Biol 1238:27–49
Bujold D, Morais DA, Gauthier C, Cote C, Caron M, Kwan T, Chen KC, Laperle J, Markovits AN, Pastinen T, Caron B, Veilleux A, Jacques PE, Bourque G (2016) The international human Epigenome consortium data portal. Cell Syst 3(5):496–499.e492
Albrecht F, List M, Bock C, Lengauer T (2016) DeepBlue epigenomic data server: programmatic data retrieval and analysis of epigenome region sets. Nucleic Acids Res 44(W1):W581–W586. https://doi.org/10.1093/nar/gkw211
Aken BL, Achuthan P, Akanni W, Amode MR, Bernsdorff F, Bhai J, Billis K, Carvalho-Silva D, Cummins C, Clapham P, Gil L, Giron CG, Gordon L, Hourlier T, Hunt SE, Janacek SH, Juettemann T, Keenan S, Laird MR, Lavidas I, Maurel T, McLaren W, Moore B, Murphy DN, Nag R, Newman V, Nuhn M, Ong CK, Parker A, Patricio M, Riat HS, Sheppard D, Sparrow H, Taylor K, Thormann A, Vullo A, Walts B, Wilder SP, Zadissa A, Kostadima M, Martin FJ, Muffato M, Perry E, Ruffier M, Staines DM, Trevanion SJ, Cunningham F, Yates A, Zerbino DR, Flicek P (2017) Ensembl 2017. Nucleic Acids Res 45(D1):D635–d642. https://doi.org/10.1093/nar/gkw1104
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Reszka, E., Zienolddiny, S. (2018). Epigenetic Basis of Circadian Rhythm Disruption in Cancer. In: Dumitrescu, R., Verma, M. (eds) Cancer Epigenetics for Precision Medicine . Methods in Molecular Biology, vol 1856. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-8751-1_10
Download citation
DOI: https://doi.org/10.1007/978-1-4939-8751-1_10
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-8750-4
Online ISBN: 978-1-4939-8751-1
eBook Packages: Springer Protocols