Abstract
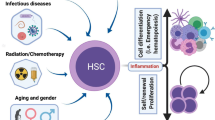
Normally, quiescent hematopoietic stem cells (HSC) rapidly enter the cell cycle following exposure to inflammatory stimuli. The analysis of HSC cell cycle activity in murine bone marrow during inflammation is often complicated by the relative rarity of HSCs and shifts in Sca-1, a key cell surface marker used to identify HSCs. Here, we report a method to analyze HSC proliferation and cell cycle distribution under inflammatory conditions. Our approach uses EdU incorporation and Ki67 staining coupled with DNA content quantification by DAPI. We also incorporate the surface marker ESAM to help minimize the potential for contaminating events that may confound analysis in the HSC compartment.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Orkin SH, Zon LI (2008) Hematopoiesis: an evolving paradigm for stem cell biology. Cell 132:631–644. doi:10.1016/j.cell.2008.01.025
Pietras EM, Warr MR, Passegué E (2011) Cell cycle regulation in hematopoietic stem cells. J Cell Biol 195:709–720. doi:10.1083/jcb.201102131
Schepers K, Campbell TB, Passegué E (2015) Normal and leukemic stem cell niches: insights and therapeutic opportunities. Cell Stem Cell 16:254–267. doi:10.1016/j.stem.2015.02.014
Rossi L, Lin KK, Boles NC et al (2012) Less is more: unveiling the functional core of hematopoietic stem cells through knockout mice. Cell Stem Cell 11:302–317. doi:10.1016/j.stem.2012.08.006
Kiel MJ, Yilmaz ÖH, Iwashita T et al (2005) SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 121:1109–1121. doi:10.1016/j.cell.2005.05.026
Wilson A, Laurenti E, Oser G et al (2008) Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 135:1118–1129. doi:10.1016/j.cell.2008.10.048
Cabezas-Wallscheid N, Klimmeck D, Hansson J et al (2014) Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell 15:507–522. doi:10.1016/j.stem.2014.07.005
Foudi A, Hochedlinger K, Van Buren D et al (2008) Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat Biotechnol 27:84–90. doi:10.1038/nbt.1517
Winkler IG, Barbier V, Wadley R et al (2010) Positioning of bone marrow hematopoietic and stromal cells relative to blood flow in vivo: serially reconstituting hematopoietic stem cells reside in distinct nonperfused niches. Blood 116:375–385. doi:10.1182/blood-2009-07-233437
Sun J, Ramos A, Chapman B et al (2014) Clonal dynamics of native haematopoiesis. Nature 514:322–327. doi:10.1038/nature13824
Busch K, Klapproth K, Barile M et al (2015) Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature 518:542–546. doi:10.1038/nature14242
Sawai CM, Babovic S, Upadhaya S et al (2016) Hematopoietic stem cells are the major source of multilineage hematopoiesis in adult animals. Immunity 45:597–609. doi:10.1016/j.immuni.2016.08.007
Pietras EM, Reynaud D, Kang Y-A et al (2015) Functionally distinct subsets of lineage-biased multipotent progenitors control blood production in normal and regenerative conditions. Cell Stem Cell 17:35–46. doi:10.1016/j.stem.2015.05.003
King KY, Goodell MA (2011) Inflammatory modulation of HSCs: viewing the HSC as a foundation for the immune response. Nat Rev Immunol 11:685–692. doi:10.1038/nri3062
Takizawa H, Boettcher S, Manz MG (2012) Demand-adapted regulation of early hematopoiesis in infection and inflammation. Blood 119:2991–3002. doi:10.1182/blood-2011-12-380113
Pietras EM, Mirantes-Barbeito C, Fong S et al (2016) Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat Cell Biol 18(6):607–618. doi:10.1038/ncb3346
Essers MAG, Offner S, Blanco-Bose WE et al (2009) IFNα activates dormant haematopoietic stem cells in vivo. Nature 458:904–908. doi:10.1038/nature07815
Ueda Y, Cain DW, Cain DW et al (2009) IL-1R type I-dependent hemopoietic stem cell proliferation is necessary for inflammatory granulopoiesis and reactive neutrophilia. J Immunol 182:6477–6484. doi:10.4049/jimmunol.0803961
Baldridge MT, King KY, Boles NC et al (2010) Quiescent haematopoietic stem cells are activated by IFN-gamma in response to chronic infection. Nature 465:793–797. doi:10.1038/nature09135
Pietras EM, Lakshminarasimhan R, Techner J-M et al (2014) Re-entry into quiescence protects hematopoietic stem cells from the killing effect of chronic exposure to type I interferons. J Exp Med 211:245–262. doi:10.1084/jem.20131043
Haas S, Hansson J, Klimmeck D et al (2015) Inflammation-induced emergency megakaryopoiesis driven by hematopoietic stem cell-like megakaryocyte progenitors. Cell Stem Cell 17:422–434. doi:10.1016/j.stem.2015.07.007
Matatall KA, Shen C-C, Challen GA, King KY (2014) Type II interferon promotes differentiation of myeloid-biased hematopoietic stem cells. Stem Cells 32:3023–3030. doi:10.1002/stem.1799
Ooi AGL, Karsunky H, Majeti R et al (2009) The adhesion molecule Esam1 is a novel hematopoietic stem cell marker. Stem Cells 27:653–661. doi:10.1634/stemcells.2008-0824
Salic A, Mitchison TJ (2008) A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci U S A 105:2415–2420. doi:10.1073/pnas.0712168105
Barbier V, Nowlan B, Lévesque J-P, Winkler IG (2012) Flow cytometry analysis of cell cycling and proliferation in mouse hematopoietic stem and progenitor cells. Methods Mol Biol 844:31–43. doi:10.1007/978-1-61779-527-5_3
Oguro H, Ding L, Morrison SJ (2013) SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell 13:102–116. doi:10.1016/j.stem.2013.05.014
Pronk CJH, Rossi DJ, Månsson R et al (2007) Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell 1:428–442. doi:10.1016/j.stem.2007.07.005
Acknowledgment
This work was supported by NIH grant K01DK98315 to E.M.P. We thank Biniam Adane and Brett Stevens for reagents and advice during the preparation of this chapter.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Science+Business Media LLC
About this protocol
Cite this protocol
Jalbert, E., Pietras, E.M. (2018). Analysis of Murine Hematopoietic Stem Cell Proliferation During Inflammation. In: Lacorazza, H. (eds) Cellular Quiescence. Methods in Molecular Biology, vol 1686. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-7371-2_14
Download citation
DOI: https://doi.org/10.1007/978-1-4939-7371-2_14
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-7370-5
Online ISBN: 978-1-4939-7371-2
eBook Packages: Springer Protocols