Abstract
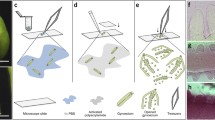
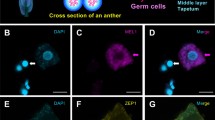
Recent advances in fluorescence-based staining of cellular compartments coupled with confocal microscopy imaging have allowed the visualization of three-dimensional (3D) structures with cellular resolution in various intact plant tissues and species. Such approaches are of particular interest for the analysis of the reproductive lineage in plants including the meiotic precursor cells deeply embedded within the ovary of the gynoecium enclosed in the flower. Yet, their relative inaccessibility and the lack of optical clarity of plant tissues prevent robust staining and imaging across several cell layers. Several whole-mount tissue staining and clearing techniques are available. One of them specifically allows staining of cellular boundaries in thick tissue samples while providing extreme optical clarity, using an acidic treatment followed by a modified Pseudo-Schiff propidium iodide (mPS-PI) method. While commonly used for Arabidopsis tissues, its application to other species like the model crop rice required protocol adaptations for obtaining robust staining that we present here. The procedure comprises six steps: (a) Material sampling; (b) Material fixation; (c) Tissue preparation; (d) Staining; (e) Sample mounting; and (d) Microscopy imaging. Particularly, we use ethanol and acetic anhydride as fixative reagents. A modified enzymatic treatment proved essential for starch degradation influencing optical clarity hence allowing acquisition of images at high resolution. This improved protocol is efficient for analyzing the megaspore mother cells in rice (Oryza sativa) ovary but is broadly applicable to other crop tissues of complex composition, without the need for tissue sectioning.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Sieber P, Gheyselinck J, Gross-Hardt R, Laux T, Grossniklaus U, Schneitz K (2004) Pattern formation during early ovule development in Arabidopsis thaliana. Dev Biol 273(2):321–334. doi:10.1016/j.ydbio.2004.05.037
Colombo L, Battaglia R, Kater MM (2008) Arabidopsis ovule development and its evolutionary conservation. Trends Plant Sci 13(8):444–450. doi:10.1016/j.tplants.2008.04.011
Cucinotta M, Colombo L, Roig-Villanova I (2014) Ovule development, a new model for lateral organ formation. Front Plant Sci 5:117. doi:10.3389/fpls.2014.00117
Truernit E, Bauby H, Dubreucq B, Grandjean O, Runions J, Barthélémy J, Palauqui JC (2008) High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of Phloem development and structure in Arabidopsis. Plant Cell 20(6):1494–1503. doi:10.1105/tpc.107.056069
Kurihara D, Mizuta Y, Sato Y, Higashiyama T (2015) ClearSee: a rapid optical clearing reagent for whole-plant fluorescence imaging. Development 142(23):4168–4179. doi:10.1242/dev.127613
Musielak TJ, Schenkel L, Kolb M, Henschen A, Bayer M (2015) A simple and versatile cell wall staining protocol to study plant reproduction. Plant Reprod 28(3–4):161–169. doi:10.1007/s00497-015-0267-1
Nguyen ST, McCurdy DW (2015) High-resolution confocal imaging of wall ingrowth deposition in plant transfer cells: semi-quantitative analysis of phloem parenchyma transfer cell development in leaf minor veins of Arabidopsis. BMC Plant Biol 15:109. doi:10.1186/s12870-015-0483-8
Yoshida S, Barbier de Reuille P, Lane B, Bassel GW, Prusinkiewicz P, Smith RS, Weijers D (2014) Genetic control of plant development by overriding a geometric division rule. Dev Cell 29(1):75–87. doi:10.1016/j.devcel.2014.02.002
Bassel GW, Smith RS (2016) Quantifying morphogenesis in plants in 4D. Curr Opin Plant Biol 29:87–94. doi:10.1016/j.pbi.2015.11.005
Coen E, Rolland-Lagan AG, Matthews M, Bangham JA, Prusinkiewicz P (2004) The genetics of geometry. Proc Natl Acad Sci U S A 101(14):4728–4735. doi:10.1073/pnas.0306308101
Fernandez R, Das P, Mirabet V, Moscardi E, Traas J, Verdeil JL, Malandain G, Godin C (2010) Imaging plant growth in 4D: robust tissue reconstruction and lineaging at cell resolution. Nat Methods 7(7):547–553. doi:10.1038/nmeth.1472
Barbier de Reuille P, Routier-Kierzkowska AL, Kierzkowski D, Bassel GW, Schüpbach T, Tauriello G, Bajpai N, Strauss S, Weber A, Kiss A, Burian A, Hofhuis H, Sapala A, Lipowczan M, Heimlicher MB, Robinson S, Bayer EM, Basler K, Koumoutsakos P, Roeder AH, Aegerter-Wilmsen T, Nakayama N, Tsiantis M, Hay A, Kwiatkowska D, Xenarios I, Kuhlemeier C, Smith RS (2015) MorphoGraphX: a platform for quantifying morphogenesis in 4D. elife 4:05864. doi:10.7554/eLife.05864
Mendocilla Sato E, Baroux C (2017) Analysis of 3D cellular organization of fixed plant tissues using a user-guided platform for image segmentation. Bio-Protocols 7(12):e2355
Stegmaier J, Amat F, Lemon WC, McDole K, Wan Y, Teodoro G, Mikut R, Keller PJ (2016) Real-time three-dimensional cell segmentation in large-scale microscopy data of developing embryos. Dev Cell 36(2):225–240. doi:10.1016/j.devcel.2015.12.028
Sankar M, Nieminen K, Ragni L, Xenarios I, Hardtke CS (2014) Automated quantitative histology reveals vascular morphodynamics during Arabidopsis hypocotyl secondary growth. elife 3:e01567. doi:10.7554/eLife.01567
Hervieux N, Dumond M, Sapala A, Routier-Kierzkowska AL, Kierzkowski D, Roeder AH, Smith RS, Boudaoud A, Hamant O (2016) A mechanical feedback restricts sepal growth and shape in Arabidopsis. Curr Biol. doi:10.1016/j.cub.2016.03.004
Summerfield RJ, Collinson ST, Ellis RH, Roberts EH, Penning de Vries FWT (1992) Photothermal responses of flowering in rice (Oryza sativa). Ann Bot 69(2):101–112. doi:10.1093/oxfordjournals.aob.a088314
Acknowledgments
This work was supported and funded by the Commission for Technology and Innovation (CTI grant 16997), the Baugarten Stiftung Zürich, the University of Zürich, and the Swiss National Science Foundation (SNF grant 31003A_149974). We acknowledge Professor Ueli Grossniklaus (UG) for scientific support and insightful discussions and technical assistants of the department for organizational support and assistance with microscopy imaging (Christoph Eichenberger, Valeria Gagliardini, Arturo Bolaños, Peter Kopf).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer Science+Business Media LLC
About this protocol
Cite this protocol
Mendocilla-Sato, E., She, W., Baroux, C. (2017). 3D Imaging of Whole-Mount Ovules at Cellular Resolution to Study Female Germline Development in Rice. In: Schmidt, A. (eds) Plant Germline Development. Methods in Molecular Biology, vol 1669. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-7286-9_3
Download citation
DOI: https://doi.org/10.1007/978-1-4939-7286-9_3
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-7285-2
Online ISBN: 978-1-4939-7286-9
eBook Packages: Springer Protocols