Abstract
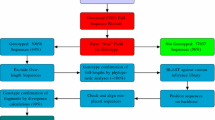
A central database to aggregate sequence information from a range of epidemiological aspects including HTLV-1 pathogenesis, origin, and evolutionary dynamic would be useful to scientists and physicians worldwide. This Chapter describes two online tools for studies related to HTLV-1, the HTLV-1 Molecular Epidemiology Database and the HTLV-1 Subtyping Tool. The HTLV-1 Molecular Epidemiology Database is a tool for sequence management and data mining which allows researchers to download sequences with clinical and demographic information. The HTLV-1 Subtyping Tool is an online software used for HTLV-1 genotyping, the algorithm consists in the alignment of a query sequence with a carefully selected set of predefined reference strains, followed by phylogenetic analysis.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Brucato N, Cassar O, Tonasso L, Tortevoye P, Migot-Nabias F, Plancoulaine S, Guitard E, Larrouy G, Gessain A, Dugoujon J-M (2010) The imprint of the Slave Trade in an African American population: mitochondrial DNA, Y chromosome and HTLV-1 analysis in the Noir Marron of French Guiana. BMC Evol Biol 10:314
Van Prooyen N, Gold H, Andresen V, Schwartz O, Jones K, Ruscetti F, Lockett S, Gudla P, Venzon D, Franchini G (2010) Human T-cell leukemia virus type 1 p8 protein increases cellular conduits and virus transmission. Proc Natl Acad Sci U S A 107:20738–20743
Filippone C, Bassot S, Betsem E, Tortevoye P, Guillotte M, Mercereau-Puijalon O, Plancoulaine S, Calattini S, Gessain A (2012) A new and frequent human T-cell leukemia virus indeterminate Western blot pattern: epidemiological determinants and PCR results in central African inhabitants. J Clin Microbiol 50:1663–1672
Mota-Miranda ACA, Barreto FK, Amarante MFC, Batista E, Monteiro-Cunha JP, Farre L, Galvão-Castro B, Alcantara LCJ (2013) Molecular characterization of HTLV-1 gp46 glycoprotein from health carriers and HAM/TSP infected individuals. Virol J 10:75
Araujo THA, Barreto FK, Luiz Carlos Júnior A, Miranda ACAM (2014) Inferences about the global scenario of human T-cell lymphotropic virus type 1 infection using data mining of viral sequences. Mem Inst Oswaldo Cruz 109:448–451
Araujo THA, Souza-Brito LI, Libin P, Deforche K, Edwards D, de Albuquerque-Junior AE, Vandamme A-M, Galvao-Castro B, Alcantara LCJ (2012) A public HTLV-1 molecular epidemiology database for sequence management and data mining. PLoS One 7:e42123
Alcantara LCJ, Cassol S, Libin P, Deforche K, Pybus OG, Van Ranst M, Galvão-Castro B, Vandamme AM, de Oliveira T (2009) A standardized framework for accurate, high-throughput genotyping of recombinant and non-recombinant viral sequences. Nucleic Acids Res 37:634–642
Wilgenbusch JC and Swofford D (2003) Inferring evolutionary trees with PAUP. Curr Protoc Bioinformatics, Chapter 6, unit 6.4
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer Science+Business Media LLC
About this protocol
Cite this protocol
Araújo, T.H.A., de Almeida Rego, F.F., Alcantara, L.C.J. (2017). Molecular Epidemiology Database for Sequence Management and Data Mining. In: Casoli, C. (eds) Human T-Lymphotropic Viruses. Methods in Molecular Biology, vol 1582. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-6872-5_2
Download citation
DOI: https://doi.org/10.1007/978-1-4939-6872-5_2
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-6870-1
Online ISBN: 978-1-4939-6872-5
eBook Packages: Springer Protocols