Abstract
Docking and quantifying the binding of small molecules to the 3D structure of a macromolecular bioregulator by computational techniques is a typical task in R&D aimed at the design and optimization of medically or otherwise active compounds. Much less known is the fact that these methods can be successfully applied for the purpose of toxicity prediction—for example, detecting a compound’s potential binding to so-called “off-targets” already at the preclinical stage. In this chapter, we provide an overview of such a computational approach, discuss its strengths and weaknesses, and include a case study—focused on natural compounds present in traditional medicines.
Access provided by CONRICYT – Journals CONACYT. Download protocol PDF
Similar content being viewed by others
Key words
- Protein-mediated toxicity
- Molecular docking
- Scoring
- Toxicity prediction
- Binding mode
- Binding affinity
- Pharmacokinetic properties
1 Introduction
Toxicology and computational chemistry are two disciplines whose synergistic combination has not been explored all too often in the past, but an ever growing importance has been witnessed. Their combination follows a concept established in rational drug design, where computational chemistry and molecular modeling are used for predicting the pharmacological activity of a small molecule—mechanistically triggered by its binding at the desired target. Analogously in toxicology, computational methods could be employed for identifying compounds leading to undesired effects as a result of their binding to relevant macromolecular targets other than the primary bioregulator—the so-called “off targets.”
2 General Concept
2.1 Pharmacokinetic Properties
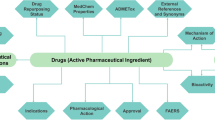
Before envisioning the computational evaluation of a compound’s ability to bind to a protein target, its availability at the site of action needs to be addressed. From the possible entry point into the human organism (e.g. transdermal, by ingestion or inhalation), the oral route has been studied in most detail [1, 2], particularly in pharmaceutical R&D, because it is the most convenient (comfortable) way of administration for the prospective patient to be treated. Knowledge gathered on the oral absorption and availability of small drug molecules is of equal importance for toxicology, because compounds associated with a harmful potential might easily reach the gastrointestinal tract (GIT) by ingestion, either intendedly (e.g. through food ingredients and additives, colorants, drugs) or unintendedly (as an undesired contaminant of any of the former).
Exploring the pharmacokinetic properties of a compound may provide hints on a compound’s specificity. In drug-design studies, it has been observed that an increasing lipophilicity of a molecule (i.e. by adding lipophilic substituents to it) might assist in improving its binding affinity, but may thereby jeopardize its specificity and decrease the ligand efficiency. Therefore, extremely lipophilic compounds (featuring a large, positive log P value) would show a non-specific interaction pattern—i.e. possibly affecting multiple targets and accumulate in adipose tissues of the body where they could persist for a prolonged period of time and possibly causing chronic adverse effects. On the other hand, hydrophilic compounds are readily filtered in the kidneys, leading to a fast clearance from the organism and, consequently, lowering the chance of triggering adverse effects.
Obtaining the most common pharmacokinetic characteristics of a given compound is quite straightforward. According to the widely accepted “Lipinski’s rule of five” [1], a compound would be likely absorbed from the GIT if its molecular weight is lower than 500, the number of hydrogen bond donors and acceptors is lower than 5 and 10, respectively, and the compound’s water–octanol partition coefficient (log P) is lower than 5. The values of the first three descriptors can be calculated by analyzing the compound’s 2D structure, while for the log P value, many trained models exist [3–7], capable to estimate the actual value by interpolation. Lipinski’s rules can be augmented with two additional rules (postulated by Veber et al.) [2] limiting the number of rotatable bonds to less than 10 and polar molecular surface area to 140 Å2.
2.2 Toxicity and Ligand Binding to Off-Targets
Toxicity and adverse effects stem from a typically non-covalent interaction (for toxicity triggered by covalently bound, i.e. reactive chemical species, please refer to ref. 8) of a small molecule with a bioregulator (receptor, enzyme, ion channel, DNA). Such an interaction can be quite unspecific, e.g. a highly lipophilic compound may be accommodated by any (at least partially) hydrophobic macromolecular cavity in order to “escape” from its interaction-wise unfavorable aqueous environment. Here, the compound’s binding would mainly be driven by desolvation effects (releasing unfavorable solvent molecules from hydrophobic cavities within a protein is beneficial for the overall binding) and weak dispersion interactions lacking any strictly preferred spatial arrangement (surface-to-surface interaction). On the other hand, a specific interaction of a small molecule with the protein target, e.g. displaying a high degree of both shape and volume complementarity to one unique protein binding site (or an allosteric or enzyme active site) with a well-defined and a thermodynamically and kinetically stable binding mode, would in addition to hydrophobic contacts likely include also several directional interactions such as salt-bridges and hydrogen bonds. In both cases, the compound’s binding to a protein may be considered as an interference with the finely tuned system of hormones, feed-back effectors, and endogenous compounds (e.g. displacing a hormone or natural substrate from the binding or active site, or transport protein, inhibiting or activating an ion channel) that would eventually perturb the physiological homeostasis within the organism and which would possibly manifest itself as adverse effects or toxicity. The impact of such effects in vivo cannot (yet) be computationally quantified with a desirable accuracy; however, the dose–response relationship would suggest that (at the given target) the more affine a compound, the more severe adverse effects or even toxicity are to be expected.
Exploring compound’s potential for protein-mediated toxicity using computational methods relies on identifying a specific non-covalent binding mode of the evaluated molecule at the macromolecular target, a concept widely used for drug design and known as molecular docking, and employing a scoring function to estimate (quantify) the binding energy.
2.3 Molecular Docking for Identifying Off-Target Binding
Molecular docking is the most convenient alternative to experimental methods directly determining the compound’s binding (e.g. in vitro assay, crystallography). Its main advantage is that it can be used also for analyzing hypothetical compounds, i.e. those not yet chemically synthesized, which allows for early screening and decision making, thus saving time and resources. The key prerequisite for application of molecular docking approach is availability of a 3D structure of the target macromolecule. This can be experimentally determined using any of the standard techniques (NMR, X-ray crystallography) or built computationally using structural information of related proteins or similar structural sub-units in homology modeling. In any case, the 3D structure, especially in the vicinity of the binding site, must be as detailed as possible with well-resolved positions of atoms in amino acids, cofactors, ligands as well as solvent (water) molecules, so that the spatial arrangement of crucial stabilizing interactions—particularly energetically prominent H-bond networks involving also water molecules—could be unambiguously determined. Ideally, several 3D structures of the target macromolecule are at hand, with different bound ligands, which can serve as templates for pre-orienting and pre-positioning of structures being docked and at the same time provide information on target’s local (e.g. amino acid side-chains flexibility) and global (e.g. backbone, loop, or large-unit rearrangement) flexibility.
When aiming at the prediction of toxicity, molecular docking is quite challenging because of typically low similarity to be expected, in terms of size, shape, and chemical composition. In addition, no template structure (bound small molecule, similar to the one of interest) might be available. In the process of lead optimization, solving the crystal structure of a lead compound bound to the target protein is therefore of utmost interest. Based on that structure, novel derivatives, typically featuring only conservative structural modifications triggering small changes in the host structure (e.g. introducing H-bond donors/acceptors or lipophilic moieties to match the binding-pocket character better), are thought to be straightforward to obtain. This implies that the new ligand’s conformation and its orientation within the binding site remains identical or, at least, similar. This fact allows to largely reduce the degrees of freedom to be explored in docking. It also simplifies the pose generation, so that even a rigid-docking protocol (keeping the macromolecule fixed) can produce reasonable results. However, when docking a compound dissimilar to any of the templates, as much structural information as possible, e.g. protein and ligand conformation, thermal displacement (B) factors, binding site shape and volume, pharmacophore assumptions, structural and displaceable solvent molecules, must be extracted from all known ligand–target structures and productively combined in order to rationalize the generation of binding modes, simultaneously decreasing the computational complexity and speeding up the docking. Random-searching algorithms (i.e. randomly modifying the ligand’s and protein’s conformation along with rotation and translation of the ligand) have theoretically a potential to identify all feasible binding modes, but due to complexity of the mathematical solution would need an enormous amount of computational time for an exhaustive sampling and therefore find only a limited use. Even thoroughly rationalized docking techniques require a rather computationally expensive geometry refinement to produce poses with reasonable interaction patterns and therefore molecular docking for predicting the off-target binding cannot be generally classified as a high-throughput method.
Binding modes generated by molecular docking allow for a mechanistic interpretation of interaction at atomic level and are of great value for further evaluation. For example, the human androgen receptor can be viewed as an anti-target as any interference with it could trigger endocrine disruption, but a compound binding to the androgen receptor identified by the docking procedure having a novel non-steroidal scaffold could serve as a basis for development of novel anabolic (agonist) or anticancer (antagonist) drugs. In case the molecule docked to the off-target would be for example a promising drug candidate, with its binding mode in hand one could modify its structure at a site that would (e.g. sterically) hinder binding to an off-target and that would be still tolerated at the desired (original) target. Such a modification might save the compound from being discarded from the development pipeline because of risk of adverse effects and even improve its selectivity and safety. In case the tested molecule would be a natural compound binding to a pharmacologically relevant target, the binding mode could indicate sites where such a structure could be simplified (e.g. removing of functional groups not involved in a favorable interaction with the target) or extended (e.g. adding a lipophilic group filling an otherwise empty part of the binding pocket) by methods of the synthetic chemistry in order to obtain a novel ligand.
2.4 Scoring Poses
While the main task of molecular docking is to identify binding modes with the most favorable ligand–target interaction energy, the scoring procedure is used to put obtained binding modes into context of a complete thermodynamic cycle, whose equilibrium is defined by the difference of free energy of the ligand and target in the unbound state and after they form a non-covalent complex. Therefore a typical scoring function, besides including enthalpic terms (electrostatic, van der Waals, H-bonding, and metal interactions), should account also for entropic terms, e.g. desolvation costs of both ligand as well as binding site at the target macromolecule, contributions stemming from solvent displacement, and penalties associated with the loss of degrees of freedom of the bound ligand and interacting amino acids in the target molecule. Entropic contributions may be calculated with a satisfactory accuracy without knowing more about dynamic properties of the interacting partners, therefore such terms are frequently approximated by summing up averaged contributions, e.g. averaged gain per displaced solvent molecules or immobilized rotatable bond, or by using empirical values [9, 10].
A scoring function might be trained in order to reproduce as closely as possible experimentally determined binding affinities of a set of compounds. However, training automatically reduces the applicability domain of a scoring function to a set of compounds similar to those in the training set. As mentioned above, the off-target binding is usually examined for compounds substantially different from those used for training (e.g. experimental binding affinities of a set of congeneric compounds from a classical medicinal chemistry lead optimization were used to train the scoring function, but a structurally dissimilar agrochemical is being evaluated), prediction based on a trained scoring function would therefore be extrapolated and very uncertain.
Despite rapid development in the field and growing complexity, there is (up-to-date) no scoring function available that would produce satisfactory results for a whole range of biologically relevant targets. Therefore, further analyses reaching beyond simple scoring, e.g. inspection of the dynamic stability of binding modes using molecular dynamics (MD) simulations or the consensus scoring employing conceptually different techniques, are highly recommended.
2.5 VirtualToxLab
The VirtualToxLab is an in silico technology for estimating the toxic potential—endocrine and metabolic disruption, some aspects of carcinogenicity and cardiotoxicity—of drugs, chemicals, and natural products [11]. The technology is based on an automated protocol that simulates and quantifies the binding of small molecules toward a series of currently 16 proteins, known or suspected to trigger adverse effects: ten nuclear receptors (androgen, estrogen α, estrogen β, glucocorticoid, liver X, mineralocorticoid, peroxisome proliferator-activated receptor γ, progesterone, thyroid α, thyroid β), four members of the cytochrome P450 enzyme family (1A2, 2C9, 2D6, 3A4), a cytosolic transcription factor (aryl hydrocarbon receptor) and a potassium ion channel (hERG). The toxic potential of a compound—its ability to trigger adverse effects—is derived from its computed binding affinities toward these very proteins (reference). The computationally demanding simulations are executed in client–server mode on a Linux cluster of the University of Basel. The graphical-user interface supports all computer platforms, allows building and uploading molecular structures, inspecting and downloading the results and, most important, rationalizing any prediction at the atomic level by interactively analyzing the binding mode of a compound with its target protein(s) in real-time 3D/4D. Access to the VirtualToxLab is available free of charge for universities, governmental agencies, regulatory bodies, and non-profit organizations.
3 Estimating the Toxic Potential of Compounds from Traditional Medicines
3.1 Rejuvenation Compounds and Traditional Medicine
We performed a study exploring compounds occurring in rejuvenating or anti-aging preparations present in various traditional medicines. The latter enjoy a large popularity especially on the Asian and African continent and whether explicable or not, are used in the maintenance of health as well as in the prevention, diagnosis, improvement, or treatment of physical and mental illnesses. Such herbal and fungal preparations contain highly species-specific secondary metabolites—compounds which might help in fighting the various symptoms of aging, such as overall weakness and decreased metabolism, reduced immunity, cognition, fertility, or muscle strength, decline in memory functions or loss of skin elasticity. Some of these symptoms can be associated with an age-related natural ligand (hormone) depletion followed by insufficient activation of associated bioregulators. For example, a low testosterone level would prevent from the androgen receptor activation and result in decreased transcription of AR-regulated genes for muscle growth. The VirtualToxLab with its target portfolio covering several nuclear receptors seems to be the right tool for screening of potential rejuvenating compounds.
The use of preparations (or single compounds isolated therefrom) recommended by traditional medicines is sometimes documented by medicinal studies—for example, antioxidants (vitamins, flavonoids) have been shown to scavenge free radical thus preventing DNA and protein from being damaged by such reactive chemical species [12], but frequently little or no evidence exists, which poses potential risks (side effects, toxicity) of “blind” usage of not properly explored and standardized preparations. On the other hand, a substantial number of modern drugs has been inspired by natural (and traditional) medicines, therefore screening such compounds by modern techniques (including in silico methods) may lead to beneficial discoveries and perhaps new drugs.
From the safety point of view, all chemical entities including natural compounds (or products of plant or animal origin containing secondary metabolites), which might occur within the human gastrointestinal tract (intended or unintended, e.g. trough food contaminants with agricultural origin) should be characterized and analyzed to the extent that we apply for pharmaceuticals.
3.2 Compound Identification and Modeling
Scientific (Pubmed, ScienceDirect) as well as general purpose (Google) electronic search engines were used along with keywords: “rejuvenat*”, “anti-ag(e)ing”, “traditional”, “medicine” to retrieve information about biological organisms and their secondary metabolites that could be associated with supposed or described biological effects. In matching publications from peer-reviewed journals, names and structure formulas of 35 unambiguously characterized secondary metabolites from seven plant and three mushroom species were identified (Table 1). Compounds with already known beneficial properties (e.g. flavonoid antioxidants, vitamins), well-researched (e.g. cardioglycosides), or acting at a different target organism (e.g. anti-infectives) were excluded from our analysis. If available, the 3D structures of the underlying compounds were retrieved from the Cambridge Structure Database (CSD) [13]. Using small-molecule crystal structure geometries as input structures when dealing with natural compounds featuring extremely complex ring systems (e.g. multiply fused and/or spiro) would seem to be appropriate as this facilitates identifying the correct ring puckering as well as correct assignment of asymmetric centers in the molecule.
The calculation of descriptors related to pharmacokinetics was performed using the Schrodinger’s QikProp program (rule-of-five violations, molecular weight [MW], polar surface area [PSA]) [14] and the VCC Lab AlogPs algorithm (Log P o/w) [7]. Finally, all structures were submitted to the VirtualToxLab for an automated simulation of the binding mode(s) and estimation of the associated affinities toward all 16 targets (cf. above). For selected ligand–target complexes, molecular dynamics simulations using the Desmond software [15] were performed to examine the dynamic stability of intermolecular interactions.
4 Results and Interpretation
4.1 Pharmacokinetic Properties
The values for the pharmacokinetic descriptors are summarized in Table 1 with favorable properties highlighted in green, potentially problematic in orange and unfavorable ones in red. With a few exceptions (e.g. panaxicol, falcarinol, and hyperforin), the studied compounds are quite rigid, lipophilic, and of low-molecular weight, thus fulfilling most of criteria defined by the Lipinski's rule-of-five. This suggests that they could be absorbed from the gastrointestinal tract after oral intake and, therefore, would be available in the systemic circulation. As a consequence of the very low PSA (<90 Å2), some of the compounds (e.g. withasomnine, carnosic acid) could even cross the blood–brain barrier and interact with bioregulators in the central nervous system. Secondary metabolites from Ginkgo biloba, despite the low number of rotatable bonds, have a rather low lipophilicity (Log P ~ 0) and a large PSA (just at, or above the limit of 140 Å2) rendering them less feasible for passive permeation and therefore less orally available. On the other hand, some compounds, e.g. panaxadiol, ganoderol A and B—due to their pronounced lipophilicity—might be quite insoluble in water and, therefore, orally available in very limited amounts, but at repeated exposure, could accumulate in the adipose tissues, where they could persist over longer periods of time.
In general, with the exception of hyperforin (which differs substantially from typical orally available molecules in molecular weight, flexibility, and lipophilicity), all studied compounds have a good chance of being absorbed after oral intake, e.g. as an extract in tonic or as a part of food. The Lipinski's rule-of-five is by no means exclusive; it solely defines descriptor ranges where there is an increased likelihood for a compound of being orally available. Therefore, a slight deviation in one or two of Lipinski’s or Veber’s descriptors from recommended values observed for a few of studied compounds does not imply that, after all, they could not be orally available.
4.2 VirtualToxLab Binding Profiles
Binding-mode hypotheses and toxic-potential values obtained by the automatic docking and scoring protocol as implemented in the VirtualToxLab are summarized in Table 2. The color intensity correlates with the predicted affinity: dark gray cells indicate hits, i.e. computationally identified complementarity of the compound with a particular binding pocket (having at least one feasible binding pose) and favorable thermodynamics of transfer from aqueous environment to the binding site. For a better understanding of the following paragraphs, selected compounds discussed in detail are depicted in Fig. 1.
Compounds with low molecular weight (e.g. anaferin, anahygrine, cuscohygrine, isopelletierine withasomnine, hispidin, and oosporein) would seem to be too small for effectively occupying the binding site of any of the screened targets. In the VirtualToxLab, these compounds do not display any significant binding affinity and, consequently, their computed toxic potential is low. No favorable binding mode could be computationally identified for the topologically complex and pronouncedly hydrophobic hyperforin. The rigid pharmacophore—the spatial arrangement of functional groups attached to complex polycyclic scaffolds—of all ginkgolides, bilobalide, Asiatic, and carnosic acid is not complementary to any binding site of the targets currently implemented in the VirtualToxLab—even though explicitly allowing for ligand flexibility and local induced-fit in our simulations. No favorable interaction with any of the 16 targets could neither be identified for (R)-ganodone, nor for (S)-ganodone. Thus, for all compounds mentioned above, no effect on the symptoms of aging could be deducted based on the results from the VirtualToxLab. This, however, does not exclude other modes of action, i.e. effects triggered through binding to targets other than nuclear receptors, enzymes of the cytochrome P450 family, and the hERG potassium channel.
Several rings as well as H-bond donor and acceptor functionalities of the essentially rigid (according to Veber “completely rigid” as terminal methyl and hydroxyl groups are not counted as rotatable in that very concept) miroestrol derivatives closely resemble the pharmacophore of the naturally occurring female hormone 17β-estradiol. This results in an increased affinity toward nuclear receptors having steroidal structures as natural ligands, especially toward α and β estrogen receptors (Table 2). Upon binding to the estrogen receptor β (ERβ; Fig. 2), some of the polar atoms of miroestrol derivatives (carbonyl, ring oxygen atom, hydroxyl group) are not involved in any favorable interaction and offer possibilities for modification, while hydroxyl groups corresponding to ones at polar ends of the estradiol should be preserved, if binding to ERβ is desired. A short molecular-dynamics simulation using the ligand–protein complex from the VirtualToxLab as the starting structure confirmed that these hydroxyl groups form stable H-bonds to the receptor (Fig. 3a). The hydroxyl group attached to the aromatic ring (corresponding to position 3 in ring A of estradiol) forms a direct H-bond with Glu305 (present during 99 % of the entire simulation time) and a water-mediated H-bond with either Arg346 (55 %) or Leu339 (15 %). The hydroxyl group mimicking the one at the 17β-position in the ring D of estradiol donates an H-bond to His475 (45 %) or Gly472 (36 %). As all three miroestrol derivatives are of comparable shape and size with estradiol, an agonistic effect is to be expected, which would seem to support the idea of administering a preparation from Pueraria mirifica containing miroestrols as estradiol mimicking molecules for relieving from symptoms associated with low estrogen levels in aging women. Obviously, instead of a rejuvenation, in men such compounds would cause an undesired feminization.
The steroidal scaffold of compounds from Withania somnifera (withanolides and similar), Panax ginseng (panaxadiol, panaxatriol, protopanaxadiol), and Ganoderma lucidum (ganoderol A and B, lucidone, ganoderenic acid A) suggests that such compounds may bind to nuclear receptors. However, most of them differ from typical natural steroidal agonists, because they feature a bulky and at least partially rigid substituent (6-membered lactone or pyran ring) at the position 17 of the cyclopentanoperhydrophenanthrene scaffold, which requires certain space for a proper accommodation and therefore could trigger induced-fit changes in the binding site leading to destabilization of the receptor structure—in this context, only partial agonistic or even antagonistic effects could be expected. In addition, the scaffold of these compounds is decorated with polar hydroxyl groups at positions different from those in natural ligands, which cannot form H-bonds with the same thermodynamic efficiency like those of latter do. Molecular-dynamics simulations of ligand–protein complexes using the highest-ranked binding pose from the VirtualToxLab as input structures showed that such hydrogen bonds have either a transient character (frequent interchange) or completely disappear early in the course of simulation (Fig. 3b), which greatly reduces their contribution toward the binding free energy (enthalpic terms). Such an unstable intermolecular interaction has been observed also for extremely flexible compounds like falcarinol and panaxicol. The computed data suggest that any potential beneficial effects of this subgroup of compounds in the context of rejuvenation might stem from weaker and not too specific binding, possibly at multiple nuclear receptors. The interactive analysis of the 4D ensemble of predicted binding modes used for scoring usually shows multiple poses with significant contributions toward the binding free energy, but with largely different orientation within the binding site accompanied by changes of side-chain conformations (local induced-fit; Fig. 4). Some compounds from the Panax species showed binding also to cytochromes (e.g. protopanaxadiol at CYP450 2D6), which might cause an enzyme inhibition and thus interfere with metabolic functions in liver cells.
At this place, we would like to point out that any outcome of an in silico screening in predictive toxicology, but especially the negative one, has to be interpreted with caution, as the applied methods and approximated model systems simply cannot provide a completely realistic answer to our scientific problem (e.g. due to a non-exhaustive conformational sampling, limited simulation time, and incomplete support for global conformational changes of target molecules, inaccuracies, or complete absence of force-field parameters).
5 Concluding Notes
In silico analyses of compounds, which are associated with rejuvenating effects based on traditional medicines, showed that a large majority of them fulfill the criteria for oral availability. This means that after ingestion they would be able to reach the systemic circulation, while some of them could even cross the blood–brain barrier and exert their effects in the central nervous system.
Computed data—in the form of binding modes at the atomic level featuring favorable H-bonding as well as hydrophobic interaction patterns with associated binding free energies obtained by state-of-the-art methodologies—seem to provide some support for potential natural hormone-mimicking effects, particularly the group of miroestrol derivatives and to a smaller extent also for some steroid-like secondary metabolites occurring in the species Withania, Panax, and Ganoderma, but also uncover the risk associated with compound's inappropriate use, lack of selectivity, and possible interference with cytochromes.
The dynamic stability of interactions between ligand and target obtained by the automated docking was explored by means of MD simulations: while a few compounds exhibit stable and well-defined binding modes to some nuclear receptors further confirming their predicted binding potential, the others form only labile interactions suggesting that the scoring function might have overestimated their binding potential.
Positive findings regarding potential biological effects described in this study highlight the importance of a proper toxicological characterization of natural compounds occurring in preparations recommended by the traditional medicine, as their uncontrolled or excessive application or unintended use might affect human health negatively.
References
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (1997) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 23:3–25
Veber DF, Johnson SR, Cheng H-Y, Smith BR, Ward KW, Kopple KD (2002) Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem 45:2615–2623
Carrupt P-A, Testa B, Gaillard P (1997) Computational approaches to lipophilicity: methods and applications. Rev Comp Chem 11:241–315
Wang R, Fu Y, Lai L (1997) A new atom-additive method for calculating partition coefficients. J Chem Inf Comput Sci 37:615–621
Kellogg GE, Abraham DJ (2000) Hydrophobicity: is LogP o/w more than the sum of its parts? Eur J Med Chem 35:651–661
Tetko IV, Tanchuk VY, Villa AE (2001) Prediction of n-octanol/water partition coefficients from PHYSPROP database using artificial neural networks and E-state indices. J Chem Inf Comput Sci 41:1407–1421
Tetko IV, Tanchuk VY (2002) Application of associative neural networks for prediction of lipophilicity in ALOGPS 2.1 program. J Chem Inf Comput Sci 42:1136–1145, http://www.vcclab.org/lab/alogps/
Kalgutkar AS (2011) Handling reactive metabolite positives in drug discovery: what has retrospective structure–toxicity analyses taught us? Chem Biol Interact 192:46–55
Spyrakis F, Kellogg GE, Amadasi A, Cozzini P (2007) Scoring functions for virtual screening. Front Drug Des Discov 3:317–379, Bentham Science Publishers Ltd., eISBN: 978-1-60805-201-1
Polgár T, Keserü GM (2007) Structure-based virtual screening. Front Drug Des Discov 3:477–502, Bentham Science Publishers Ltd., eISBN: 978-1-60805-201-1
Vedani A, Dobler M, Hu Z, Smieško M (2015) OpenVirtualToxLab—a platform for generating and exchanging in silico toxicity data. Toxicol Lett 232:519–532, available at http://www.virtualtoxlab.org/
Kumar S, Pandey K (2013) Chemistry and biological activities of flavonoids: an overview. Sci World J 2013:162750. doi:10.1155/2013/162750
Allen FH (2002) The Cambridge Structural Database: a quarter of a million crystal structures and rising. Acta Crystallogr B 58:380–388
QikProp, version 4.0 (2014) Schrödinger, LLC, New York
Desmond Molecular Dynamics System, version 3.8 (2014) D. E. Shaw Research, New York. Maestro-Desmond Interoperability Tools, version 3.8 (2014) Schrödinger, New York
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer Science+Business Media New York
About this protocol
Cite this protocol
Smieško, M., Vedani, A. (2016). VirtualToxLab: Exploring the Toxic Potential of Rejuvenating Substances Found in Traditional Medicines. In: Benfenati, E. (eds) In Silico Methods for Predicting Drug Toxicity. Methods in Molecular Biology, vol 1425. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-3609-0_7
Download citation
DOI: https://doi.org/10.1007/978-1-4939-3609-0_7
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-3607-6
Online ISBN: 978-1-4939-3609-0
eBook Packages: Springer Protocols