Abstract
Photoreceptors are found in all kingdoms of life and mediate crucial responses to environmental challenges. Nature has evolved various types of photoresponsive protein structures with different chromophores and signaling concepts for their given purpose. The abundance of these signaling proteins as found nowadays by (meta-)genomic screens enriched the palette of optogenetic tools significantly. In addition, molecular insights into signal transduction mechanisms and design principles from biophysical studies and from structural and mechanistic comparison of homologous proteins opened seemingly unlimited possibilities for customizing the naturally occurring proteins for a given optogenetic task. Here, a brief overview on the photoreceptor concepts already established as optogenetic tools in natural or engineered form, their photochemistry and their signaling/design principles is given. Finally, so far not regarded photosensitive modules and protein architectures with potential for optogenetic application are described.
Access provided by CONRICYT – Journals CONACYT. Download protocol PDF
Similar content being viewed by others
Key words
1 Natural Photoreceptors
1.1 Chromophores
Naturally occurring photoreceptors cover the spectral range from the UV-B to the near-IR (Fig. 1). For this purpose, different chromophores derived from metabolic compounds and enzyme cofactors are utilized and their electronic properties are fine-tuned by the protein environment. Until lately the retinal cofactor of visual and nonvisual opsins has been considered the most versatile chromophore spanning from the near UV regions (~360 nm) in short wavelength sensitive rhodopsins (SWS) [1] to ~700 nm in one of the mantis shrimp rhodopsins [2]. With the discovery algal phytochromes [3] and especially of a novel type of linear tetrapyrrole binding photoreceptor, cyanobacteriochrome , which expand the range of tetrapyrrole chromophores to cover all of the visible spectrum and the near UV, tetrapyrroles outperform retinal in spectral coverage [3, 4]. In the blue to near-UV region, flavin binding photoreceptors—LOV (light–oxygen–voltage), BLUF (blue light sensors using FAD), and cryptochrome photoreceptors—are prominent and cover a large spectral region with their first two electronic transitions in the oxidized form but are rather limited in color tuning [5]. However, in different reduced states, semiquinone or fully reduced form, flavins have been described to respond to green/yellow or UV-A, respectively [6–8]. In the UV-B region, a recently described photoreceptor concept, UVR8 , uses no cofactor at all and employs the UV absorbing nature of tryptophan side chains as a proteinogenic chromophore [9]. In addition, a few special cases exist like the Xanthopsins that employ p-coumaric acid as a blue light absorbing chromophore [10] or the just recently discovered cobalamin binding photoreceptors absorbing in the green to red wavelength region [11].
The photochemical properties of most of these chromophores in isolated and protein-bound form are well known. The key biophysical determinants for photoactivation are the quantum efficiency of signaling state generation and its deactivation reaction in the dark (Table 1). These properties directly affect the net biological signal/effect. The observed photoinduced reactions that lead to a signaling active conformation are isomerizations (retinal , tetrapyrrole , p-coumaric acid ), proton coupled electron transfer (tryptophan, flavin), and chemical/redox reactions (flavin). The general photochemistry of the chromophores can be readily probed with state-of-the art time-resolved spectroscopic techniques [12, 13]; however, the interplay with the protein and the relevant structural changes in signal transduction are in most cases more difficult to address. The latter are essential for the feasibility of a potential optogenetic tool, since the light-dependent modulation of the biological signal is not only determined by the quantum yield of the phototransformation of the chromophore itself but rather by the stability of the resulting signaling state and its impact on the protein structure/the extent of modulation of effector activity . In addition to the photoactivation properties, thermal/dark noise of the given photosensor as discussed in GPCR signaling is a relevant aspect [14].
Secondary aspects are of course phototoxicity by photoinduced generation of reactive oxygen species (ROS) byproducts in the photoactivation process itself or by absorption in the cell/tissue that have to be considered for application and naturally depend on the energy required for sufficient optical stimulation . The latter is especially crucial in tissue or animals, where penetration depth is limiting. In such cases, long-wavelength excitation is desirable to minimize diffusion/scattering of the stimulating light. Moreover, energy dissipation from the receptor protein and the tissue itself are crucial parameters for design and evaluation of optogenetic experiments [15].
The ultimate requirement, however, is the bioavailability of the cofactor in the investigated cell type . While proteinogenic cofactors and flavins are ubiquitous in nature, the availability retinal and tetrapyrrole based cofactors may be limiting for the given application. Moreover, spectral orthogonality may have to be considered for applications where multiple selective stimuli are desired.
1.2 Photosensitive Modules/Chromophore Binding Domains
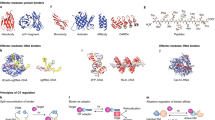
Representative crystal structural models of the binding domains for the above-described chromophores are shown in Fig. 2. While different architectures are found to bind flavins (PAS: Per–Arnt–Sim, BLUF: Blue Light sensors Using FAD, Cryptochrome/Photolyase), also some domain classes (e.g., PAS) are versatile in binding different chromophores (flavins, p-coumaric acid, tetrapyrroles ). As mentioned above, the potential for optogenetics of these classes naturally depends on the given biophysical properties (Table 1). The small modular photoreceptors are usually more convenient to deliver into the given cell or organism, but most of these small molecule binding receptors feature a low molar absorption cross section, which makes them generally less attractive for efficient photostimulation. Another important parameter is the quantum yield of phototransformation that illustrates the efficiency of the formation of the metastable signaling state. This is especially crucial in the case of the rhodopsin pumps where the phototransformation is stochiometrically related to the number of translocated ions. However, as described above the apparent modulation of the biological activity in most other photoreceptors depends strongly on how well regulated the cognate effector domain is and how strongly its activity becomes enhanced in the activated state. Therefore, even a photoreceptor with apparent low photochemical transformation efficiency (e.g., phytochrome) can be a potent optogenetic tool. Finally, the interaction with the given natural metabolism has to be considered as it might strongly lower the apparent activation compared to in vitro studies.
Photoresponsive modules found in nature: Xanthopsins , LOV domains and phyto/-cyanobacteriochromes employ the PAS fold to assemble the light-sensitive module. Cyanobacteriochromes lack the PHY domain in contrast to phytochromes. BLUF domains use a ferredoxin-like fold to bind a flavin chromophore. Cryptochromes are structurally related to photolyases and bind FAD and an antenna cofactor (not shown). UVR8 belongs to the family of WD40 repeat proteins with a characteristic β-propeller structure. Pumping and channelrhodopsins belong to the greater family of seven transmembrane proteins and more specifically to the microbial rhodopsins. Structural data (PDB ID) used for illustration: Xanthopsin (1NWZ), AsLOV2 (2V1A), SyPixD (2HFO chain A), AtCry (4GU5), UVR8 (4D9S), C1C2 (3UG9), bacteriophytochrome (4O0P)
1.3 Signal Transduction Concepts
The above described photoreceptor classes use various concepts to transform the local structural changes of the chromophore and its protein surrounding into a biological signal and can be roughly categorized in four categories (Fig. 3). In case of the channelrhodopsins and pumping rhodopsins, these processes are intricately entangled as their activity, channeling, or ion pumping, directly involves the chromophore. Similarly, the UV-B induced monomerization of UVR8 is accomplished by direct interaction of the excited chromophore with the cross dimer salt-bridges [16, 17]. In the remaining photoreceptor classes and the sensory rhodopsins, the chromophore is in most cases located distal from the biological effector part, which is especially obvious for modular photoreceptor designs. The signaling concepts of such receptors can be classified into directly allosteric or allosterically induced uncaging and association/dissociation mechanisms [18].
The so-to-speak integral and modular photoreceptors provide different opportunities and limitations for optogenetic application and customization, accordingly. Integral photoreceptors are limited for optogenetic application solely due to their characteristic dark/thermal noise which results in more or less pronounced background signaling activity [14]. In this sense, most modular photoreceptor designs are more prone to background activity since the photosensory domain only modulates the activity of the effector domain and the dark activity thus depends strongly on the tightness of regulation by the photosensory domain [18]. The possibility to alter the principal functionality of an integral receptor is, however, very limited while modular photoreceptors are found in different sensor/effector combinations covering a wide range of biological activities already in their natural form. In addition, many photosensory modules elicit rather large scale structural changes like unfolding or (de-)oligomerization, which can be easily exploited in order to recruit or release other biomolecules.
1.4 Natural Resources of Light-Responsive Modules and Signaling Architectures
In contrast to the early times of genome analysis, the technological development in sequence analysis in combination with computational resources for automatic annotation available nowadays provides access to a vast and everyday increasing amount of information on the primary structure and, thus, on the function of proteins with known homologies. Especially the rise of metagenomics that does not rely on monoclonal samples provides access to diverse ensembles of organisms and their genomes and further increased the abundance of sequence information [19–21].
Currently, there are more than 3000 sequences available for the bacteriorhodopsin-like protein family PF01036 (http://pfam.xfam.org/family/PF01036) that feature ion pumps, ion channels, and sensory proteins. The greater family of seven transmembrane receptors (http://pfam.xfam.org/family/PF00001) with more than 58,000 sequences, covers about 8000 protein sequences of visual and nonvisual rhodopsins (http://www.ebi.ac.uk/interpro/entry/IPR001760). Even more remarkable is the greater family of PAS domains with more than 26,000 sequences (http://pfam.xfam.org/family/PF13426) that includes the LOV photosensory domains, Xanthopsins and phytochrome architectures. Less than 5 % of these sequences are found as single PAS domain containing proteins while the majority is arranged in a modular fashion in various architectures and with different effector domains. Given the high homology of PAS domains this abundance of structures provides a large number of naturally optimized architectures that can possibly be rendered light sensitive by replacing the given PAS domain with a suitable photosensitive domain, e.g., LOV. Similarly impressive are the phytochrome family (http://pfam.xfam.org/family/PF00360) that currently encompasses about 6300 sequences containing PAS–GAF(cGMP phosphodiesterase , adenylate cyclase , FhlA)–PHY(phytochrome-specific GAF related) modules and the BLUF family with about 3900 sequences (http://pfam.xfam.org/family/PF04940). Both classes feature a similar diversity in attached effector domains. The family of the cryptochromes (http://pfam.xfam.org/family/PF12546) currently features only 157 sequences, probably owing to their occurrence mainly in higher eukaryotic organisms for which less sequence information is available. The structurally closely related photolyase family (http://pfam.xfam.org/family/PF00875), however, features about 15,000 sequences.
The current knowledge on photosensitive modules and receptor architecture thus allows us to extract homologous proteins from genomic or metagenomic data, predict to some extent their properties, screen for desired traits and learn about modular architecture that can be exploited for the rational design of artificial photoreceptors. However, proteins with novel photo-activation concepts cannot be easily discovered in this way and still require thorough photo- and cell biological research. A few recent examples are given below.
2 Engineering of Photoreceptors and Artificial Signal Transduction Pathways
2.1 Photoreceptor Engineering
Besides directly harnessing the possibilities provided by nature itself, many photoreceptor functionalities have been extensively engineered. We only focus on a few selected examples, as concise review articles have been published recently (e.g., [18] and chapters 27 in this book). One of the most prominent examples is channelrhodopsin, which has been altered in its activation and deactivation properties, spectral sensitivity, and even ion selectivity [22–25]. Except for PACs, which have also been applied successfully in their natural form [26–29], most modular photoreceptors used in optogenetics have been cut down to the photosensitive module and fused with different interaction partners [18, 30]. The reason for this may be due to the related dark activity or unsuitable signaling activity of the given natural system.
For example, the light-modulated transcription factor Aureochrome found in yellow algae and diatoms at first glance seems like a perfect minimal system to regulate transcription in heterologous systems [31]. However, the short recognition sequence of the bZIP (Basic Leucine Zipper ) domain and the ratio of light/dark activity prevent an efficient transfer of these systems into heterologous environments [32]. In its natural context probably other transcription effectors or regulatory elements are engaged to facilitate a physiologically sufficient light-induced change in activity. Therefore, a more efficient approach was to employ a hybrid approach using a (heterologous) LOV domain that undergoes light-induced dimerization, fused to a homologous DNA binding and a transactivator domain of the target cell type . Accordingly, light-induced binding of the homologous fusion protein to a specific sequence and a subsequent expression induction is facilitated [33]. Similarly, the naturally occurring LOV–HTH (helix–turn–helix) protein EL222 that naturally uses light-induced dimerization was fused with a transactivator domain, most likely to enhance optogenetic activation in the heterologous system [34]. Both systems have been applied successfully but differ in their dynamic range, mainly due to the lifetime of the signaling state of the given LOV domain.
In other artificial fusion constructs several modes of activation, uncaging, (de-)oligomerization, and even allostery directly have been efficiently employed. The most prominent uncaging scenario originates from the plant phototropins . Photoactivation of the LOV2 domain induces dissociation and unfolding of a short C-terminal helix [35] attached to the LOV-domain core and can be efficiently used, e.g., for uncaging of a RAC GTPase [36], a peptide toxin [37] or a cation channel [38]. Partial light-induced unfolding of a PAS domain is also witnessed in the photoactive yellow protein (PYP) and has been exploited for controlling protein/protein interactions [39].
Some naturally occurring LOV domains show light-induced dimerization that was successfully employed by fusion to DNA-binding motifs as described above [40, 41]. PYP in contrast can be engineered to feature such a photoinduced transformation [42]. Especially powerful also proved to be light-dependent heterodimerization of the Arabidopsis photoreceptors cryptochrome 2 (Arabidopsis) and CIB1 (cryptochrome-interacting basic-helix–loop–helix) [43] or phytochrome B and PIF (phytochrome-interacting factors) proteins [44], that was efficiently used to recruit and release proteins. Cryptochrome 2 (CRY2) also features homodimerization /clustering that was optimized and applied for spatial accumulation of proteins [45–47]. Similarly powerful proved to be the UV-B induced dissociation reaction of UVR8 , that was used to drive expression or nuclear localization using the natural interaction partner COP1 (CONSTITUTIVELY PHOTOMORPHOGENIC 1) [48]. Despite its large size and slow dark reversion, the specific excitation with UV-B light, which is unbiased by the observation light source in light microscopic experiments, and the missing need for an exogenous cofactor makes UVR8 especially attractive as a fully genetically encoded photoswitch.
The above-described engineering approaches employ secondary effects of light-induced allosteric changes. Engineering allostery directly, however, is significantly more difficult due to the lack of knowledge on the molecular prerequisites in most cases. In the successful cases, careful homologies between different sensor/effector constructs have been investigated and applied in the construction of the artificial protein [49–53].
2.2 Orthogonal Signal Engineering
Besides changing the photosensory properties of a given photoreceptor or de- and reconstructing of sensor/effector combinations, nature also provides us with elegant ways of introducing complete artificial signaling cascades into a heterologous system. These approaches are especially interesting since highly orthogonal systems can be designed, that ideally prevent interference with the natural metabolism in a given cell. In this regard, especially modular photoreceptor architectures are appealing as an effector domain with a suitable activity can be chosen or engineered.
The most versatile approach in this regard is the optogenetic regulation of expression. An elegant example is the hybrid light-activated histidine kinase YF1, that has been engineered by exchanging a redox sensitive PAS domain from the oxygen activated histidine kinase FixL from Bradyrhizobium japonicum for a LOV domain from Bacillus subtilis [49, 54]. Together with its downstream phosphorylation target FixK and the well-described activation of its cognate FixK2 promotor an orthogonal light-activated expression systems for E. coli was established. Due to the high specificity of such bacterial, two component systems, a cross talk with endogenous signaling cascades is unlikely.
Similarly, it is conceivable that non-native second messenger modules may be used for a given optogenetic task, e.g., to regulate the activity of a protein. At first glance, promising candidates seemed to be represented by dicyclic nucleotides, which regulate various processes in bacteria and corresponding (light-activated) cyclases and phosphodiesterases have been already described und functionally investigated [55–57]. However, only recently receptor proteins for c-di-GMP, c-di-AMP, and cGAMP have been discovered and functionally described in eukaryotes and even in mammals [58]. Accordingly, dicyclic nucleotides cannot be considered orthogonal second messengers in eukaryotic systems.
A more promising approach for orthogonality may therefore be the integration with chemical biology. Using so-called optochemical genetics it is possible to render an endogenous compound selectively light responsive with only minimal perturbance of the natural system [59, 60]. Moreover, it is possible to investigate the effect of light on the given cell independently of optical stimulation as in most cases the chromophore /photoactive ligand can be added independently of the expressed modified receptor. Similarly, it should be possible to generate non-natural enzyme activities that produce, in a light-responsive manner, artificial second messengers that are recognized by a cognate, non-natural designed receptor.
3 Potential Future Optogenetic Tools Found in Nature
The recent advances in optogenetics and optochemical genetics demonstrate that the combination of natural resources with knowledge driven molecular engineering provides seemingly endless opportunities for synthetic biology and biotechnology. Still, completely new photoreceptor principles are likely to be discovered and will add to the palette of the optogenetic construction kit in the future.
Apart from the so far described and successfully applied opsin based tools several new microbial and also some longer known opsins represent very promising candidates for optogenetics. The histidine kinase rhodopsins discovered in green algae , for example, show a unique multidomain architecture [61]. Directly fused to the transmembrane rhodopsin part are several soluble domains that constitute a complete phosphotransfer cascade on a single polypeptide. The terminal effector domain is a GTP cyclase and thus a promising natural light-activated cGMP synthase. Its function has yet to be demonstrated but the signaling network given by the protein architecture suggests an intricately regulated cyclase activity that offers many levers for optimization/adaptation in an optogenetic context. Especially intriguing is furthermore the bimodal nature of the rhodopsin photosensory domain of HKR1 that can be switched permanently between two states by UV and blue light, respectively. In that sense, also the long known non-visual rhodopsins like melanopsins are highly appealing as they also represent bimodal switches [62] and can be used to drive various G-protein signaling cascades. Melanopsins have recently been applied in modified forms in optogenetic assays for vision restoration [63] and controlling cardiomyocyte activity [64].
Another interesting modular photoreceptor candidate is a recently discovered directly rhodopsin coupled guanylyl cyclase identified in the fungus Blastocladiella emersonii [65]. The protein may be considered as a short-cut Gαs GPCR that usually requires G-proteins to stimulate cyclase activity. Its potential for optogenetics has been recently demonstrated [66, 67]. In contrast to the BLUF regulated photoactivated cyclases, the protein is intrinsically membrane localized and thus enables subcellularly localized stimulation. It is very likely that the cGMP synthesis activity can be adapted to cAMP synthesis as shown vice versa for the BLUF activated PACs [68] and would add a novel membrane localized PAC to the optogenetic toolbox.
A noteworthy subclass of microbial rhodopsins, the xanthorhodopsins [69], employ carotenoid antennas (salinixanthin ) to extend the s pectral sensitivity to facilitate light-induced proton pumping. This concept is highly interesting for color and sensitivity tuning of potentially any kind of rhodopsin, but has yet to be explored in optogenetics. Another carotenoid employing photoreceptor is the orange carotenoid protein (OCP), which has been found to regulate non-photochemical quenching in cyanobacteria. The carotenoid chromophore (3′-hydroxyechinenone) enables photochromic switching between an orange and a red form [70]. A model involving a large scale translocation of the carotenoid and accompanying structural changes at the C-terminus has been recently proposed [71], which would enable the protein to interact in a regulatory manner with the phycobilisome [72]. OCP belongs to a smaller protein family of about 216 sequences containing a cyanobacteria-specific N-terminal domain (http://pfam.xfam.org/family/PF09150). The remaining protein belongs to the class of Nuclear transport factor 2 (NTF2) domain (~6200 sequences; http://pfam.xfam.org/family/PF02136). So far this photoactivation concept has not been applied in an optogenetic context, but clearly provides interesting novel starting points for the design of optogenetic tools regarding new chromophores and specific light-induced structural changes.
Similarly to the photochemical diversity of retinal binding photoreceptors, also novel photochemical traits of the tetrapyrrole binding receptors have been discovered recently. Although phytochromes already represent near-IR/far-IR bimodal switches, the cyanobacteriochromes can be tuned over a broad range in the visible spectrum and feature well separated spectra of both states. Moreover, these proteins do not require the PHY domain that is needed for efficient photoswitching in phytochromes. These properties are clearly attractive for optogenetics and a widespread use of cyanobacteriochrome in optogenetic application is expected in the future. In addition, novel insights into the monomerization/dimerization prerequisites for phytochromes have been found recently. Inspired by the search for monomeric phytochrome based infrared fluorescent proteins [73], a photochromic monomer/dimer switchable phytochrome module has been described and clearly has a high potential for building tightly regulated bimodal optogenetic mono-/dimerizers [74].
In the first optogenetic experiments, complete visual rhodopsin signaling cascades [75] and modified rhodopsins [76–78] have been employed to stimulate G-protein induced responses. Because of the abundance of GPCRs in higher animals and their relevance for disease and metabolic disorders, GPCRs are naturally attractive targets for optogenetic studies or even therapeutics. A noteworthy completely new type of photoactivatable GPCRs has been found to be responsible for the photoavoidance of Caenorhabditis elegans and larvae of Drosophila melanogaste r [79–82]. Apparently, these gustatory GPCR paralogs (family of 7TM chemosensory receptors; http://pfam.xfam.org/family/PF08395; currently 1900 sequences) facilitate the UV-induced photophobic behavior via different G-protein cascades. LITE-1 found in C. elegans mediates photoavoidance via Gi/o proteins that stimulate the activity of membrane associated guanylyl cyclases which in turn affect CNG channels [80]. Although action spectra of the LITE-1 mediated photoresponse have been described, the nature of its chromophore is completely unclear. From the crude action spectra recorded by behavioral experiments showing a maximum in the UV-A region at around 350 nm but also a significant response at up to 450 nm, many potential chromophores can be discussed (see Fig. 1). Besides using classical prosthetic chromophores LITE-1 may in principle also transiently bind small molecules that have been chemically altered by short wavelength /high-energy light. The discussion whether LITE-1 and the related Gur-3 may also be activated by ROS as a photoinduced by-product is discussed controversially [83]. In D. melanogaster, the close homolog Grb28 (isoform B) also employs G-protein mediated signaling, but the classification of the interacting G-protein remains unknown. In contrast to LITE-1, the observed Ca2+ photocurrents are not mediated by CNG channels. Instead, the transient receptor potential (TRP) channel TrpA1 was identified to be responsible. Clearly, this class of proteins features promising candidates to stimulate various G-protein related activities in optogenetic approaches.
Another completely new class of soluble photoreceptor that was found in Myxococcus xanthus uses a closed ring tetrapyrrole cofactor, Cobalamin (coenzyme B12), and is the first appearance of this chromophore in the photoreceptor world [11]. CarH requires coenzyme B12 for light-dependent gene repressor activity by oligomerization and subsequent DNA binding. Upon illumination the cofactor is photolysed and the oligomer dismantles and dissociates from its cognate operator. The underlying photodynamics and crystal structures in dark and light-activated states have been recently described [84]. Therefore, a solid basis for application of CarH as a novel optogenetic tool has been already established.
4 Concluding Remarks
Up to now nature has provided us with a vast toolbox for optogenetic application and most likely has further so far unknown photoactivation concepts in its repertoire. Moreover, even for well-known photoreceptor concepts novel photochemical traits or signal transduction scenarios are discovered constantly. Most of these proteins provide new opportunities for optogenetic application, either to be harnessed from nature directly or in the sense that we learn about different molecular solutions for photoreception that can be applied in the rational design of new optogenetic tools. Furthermore, combination of synthetic biology with chemical biology is expected to add significantly to the optogenetic toolbox.
References
Hauser FE, van Hazel I, Chang BSW (2014) Spectral tuning in vertebrate short wavelength-sensitive 1 (SWS1) visual pigments: can wavelength sensitivity be inferred from sequence data? J Exp Zool B 322(7):529–539
Thoen HH, How MJ, Chiou TH, Marshall J (2014) A different form of color vision in mantis shrimp. Science 343(6169):411–413
Rockwell NC, Duanmu D, Martin SS, Bachy C, Price DC, Bhattacharya D et al (2014) Eukaryotic algal phytochromes span the visible spectrum. Proc Natl Acad Sci U S A 111(10):3871–3876
Ishizuka T, Shimada T, Okajima K, Yoshihara S, Ochiai Y, Katayama M et al (2006) Characterization of cyanobacteriochrome TePixJ from a thermophilic cyanobacterium Thermosynechococcus elongatus strain BP-1. Plant Cell Physiol 47(9):1251–1261
Losi A, Gärtner W (2012) The evolution of flavin-binding photoreceptors: an ancient chromophore serving trendy blue-light sensors. Annu Rev Plant Biol 63:49–72
Bouly JP, Schleicher E, Dionisio-Sese M, Vandenbussche F, Van Der Straeten D, Bakrim N et al (2007) Cryptochrome blue light photoreceptors are activated through interconversion of flavin redox states. J Biol Chem 282(13):9383–9391
Carell T, Burgdorf LT, Kundu LM, Cichon M (2001) The mechanism of action of DNA photolyases. Curr Opin Chem Biol 5(5):491–498
Beel B, Prager K, Spexard M, Sasso S, Weiss D, Muller N et al (2012) A flavin binding cryptochrome photoreceptor responds to both blue and red light in Chlamydomonas reinhardtii. Plant Cell 24(7):2992–3008
Jenkins GI (2014) The UV-B photoreceptor UVR8: from structure to physiology. Plant Cell 26(1):21–37
Meyer TE, Kyndt JA, Memmi S, Moser T, Colon-Acevedo B, Devreese B et al (2012) The growing family of photoactive yellow proteins and their presumed functional roles. Photochem Photobiol Sci 11(10):1495–1514
Ortiz-Guerrero JM, Polanco MC, Murillo FJ, Padmanabhan S, Elias-Arnanz M (2011) Light-dependent gene regulation by a coenzyme B12-based photoreceptor. Proc Natl Acad Sci U S A 108(18):7565–7570
Mathes T, van Stokkum IHM, Kennis JTM (2014) Photoactivation mechanisms of flavin-binding photoreceptors revealed through ultrafast spectroscopy and global analysis methods. Flavins Flavoproteins Methods Protocols 1146:401–442
Kennis JTM, Mathes T (2013) Molecular eyes: proteins that transform light into biological information. Interface Focus 3(5):20130005
Barlow RB, Birge RR, Kaplan E, Tallent JR (1993) On the molecular-origin of photoreceptor noise. Nature 366(6450):64–66
Stujenske JM, Spellman T, Gordon JA (2015) Modeling the spatiotemporal dynamics of light and heat propagation for in vivo optogenetics. Cell Rep 12(3):525–534
Mathes T, Heilmann M, Pandit A, Zhu J, Ravensbergen J, Kloz M et al (2015) Proton-coupled electron transfer constitutes the photoactivation mechanism of the plant photoreceptor UVR8. J Am Chem Soc 137(25):8113–8120
Heilmann M, Christie JM, Kennis JT, Jenkins GI, Mathes T (2015) Photoinduced transformation of UVR8 monitored by vibrational and fluorescence spectroscopy. Photochem Photobiol Sci 14(2):252–257
Ziegler T, Möglich A (2015) Photoreceptor engineering. Front Mol Biosci 2:30
Klapoetke NC, Murata Y, Kim SS, Pulver SR, Birdsey-Benson A, Cho YK et al (2014) Independent optical excitation of distinct neural populations. Nat Methods 11(3):338–346
Pathak GP, Losi A, Gärtner W (2011) Metagenome-based screening reveals worldwide distribution of LOV-domain proteins. Photochem Photobiol 88(1):107–118
Pathak GP, Ehrenreich A, Losi A, Streit WR, Gärtner W (2009) Novel blue light-sensitive proteins from a metagenomic approach. Environ Microbiol 11(9):2388–2399
Prigge M, Schneider F, Tsunoda SP, Shilyansky C, Wietek J, Deisseroth K et al (2012) Color-tuned channelrhodopsins for multiwavelength optogenetics. J Biol Chem 287(38):31804–31812
Wietek J, Wiegert JS, Adeishvili N, Schneider F, Watanabe H, Tsunoda SP et al (2014) Conversion of channelrhodopsin into a light-gated chloride channel. Science 344(6182):409–412
Kleinlogel S, Feldbauer K, Dempski RE, Fotis H, Wood PG, Bamann C et al (2011) Ultra light-sensitive and fast neuronal activation with the Ca(2)+-permeable channelrhodopsin CatCh. Nat Neurosci 14(4):513–518
Berndt A, Yizhar O, Gunaydin LA, Hegemann P, Deisseroth K (2009) Bi-stable neural state switches. Nat Neurosci 12(2):229–234
Weissenberger S, Schultheis C, Liewald JF, Erbguth K, Nagel G, Gottschalk A (2011) PACalpha--an optogenetic tool for in vivo manipulation of cellular cAMP levels, neurotransmitter release, and behavior in Caenorhabditis elegans. J Neurochem 116(4):616–625
Stierl M, Stumpf P, Udwari D, Gueta R, Hagedorn R, Losi A et al (2011) Light modulation of cellular cAMP by a small bacterial photoactivated adenylyl cyclase, bPAC, of the soil bacterium Beggiatoa. J Biol Chem 286(2):1181–1188
Schröder-Lang S, Schwarzel M, Seifert R, Strunker T, Kateriya S, Looser J et al (2007) Fast manipulation of cellular cAMP level by light in vivo. Nat Methods 4(1):39–42
Chen ZH, Raffelberg S, Losi A, Schaap P, Gärtner W (2014) A cyanobacterial light activated adenylyl cyclase partially restores development of a Dictyostelium discoideum, adenylyl cyclase a null mutant. J Biotechnol 191:246–249
Möglich A, Moffat K (2010) Engineered photoreceptors as novel optogenetic tools. Photochem Photobiol Sci 9(10):1286–1300
Takahashi F, Yamagata D, Ishikawa M, Fukamatsu Y, Ogura Y, Kasahara M et al (2007) AUREOCHROME, a photoreceptor required for photomorphogenesis in stramenopiles. Proc Natl Acad Sci U S A 104(49):19625–19630
Hisatomi O, Nakatani Y, Takeuchi K, Takahashi F, Kataoka H (2014) Blue light-induced dimerization of monomeric aureochrome-1 enhances its affinity for the target sequence. J Biol Chem 289(25):17379–17391
Wang X, Chen XJ, Yang Y (2012) Spatiotemporal control of gene expression by a light-switchable transgene system. Nat Methods 9(3):266–269
Motta-Mena LB, Reade A, Mallory MJ, Glantz S, Weiner OD, Lynch KW et al (2014) An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat Chem Biol 10(3):196–202
Harper SM, Neil LC, Gardner KH (2003) Structural basis of a phototropin light switch. Science 301(5639):1541–1544
Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B et al (2009) A genetically encoded photoactivatable Rac controls the motility of living cells. Nature 461(7260):104–108
Schmidt D, Tillberg PW, Chen F, Boyden ES (2014) A fully genetically encoded protein architecture for optical control of peptide ligand concentration. Nat Commun 5:3019
Cosentino C, Alberio L, Gazzarrini S, Aquila M, Romano E, Cermenati S et al (2015) Optogenetics. Engineering of a light-gated potassium channel. Science 348(6235):707–710
Morgan SA, Al-Abdul-Wahid S, Woolley GA (2010) Structure-based design of a photocontrolled DNA binding protein. J Mol Biol 399(1):94–112
Strickland D, Lin Y, Wagner E, Hope CM, Zayner J, Antoniou C et al (2012) TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat Methods 9(4):379–384
Pathak GP, Strickland D, Vrana JD, Tucker CL (2014) Benchmarking of optical dimerizer systems. ACS Synth Biol 3(11):832–838
Reis JM, Burns DC, Woolley GA (2014) Optical control of protein-protein interactions via blue light-induced domain swapping. Biochemistry 53(30):5008–5016
Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL (2010) Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods 7(12):973–975, advance online publication
Toettcher JE, Gong D, Lim WA, Weiner OD (2011) Light-based feedback for controlling intracellular signaling dynamics. Nat Methods 8(10):837–839
Bugaj LJ, Choksi AT, Mesuda CK, Kane RS, Schaffer DV (2013) Optogenetic protein clustering and signaling activation in mammalian cells. Nat Methods 10(3):249–252
Taslimi A, Vrana JD, Chen D, Borinskaya S, Mayer BJ, Kennedy MJ et al (2014) An optimized optogenetic clustering tool for probing protein interaction and function. Nat Commun 5:4925
Che DL, Duan L, Zhang K, Cui B (2015) The dual characteristics of light-induced cryptochrome 2, homo-oligomerization and heterodimerization, for optogenetic manipulation in mammalian cells. ACS Synth Biol 4(10):1124–1135
Kianianmomeni A (2015) UVB-based optogenetic tools. Trends Biotechnol 33(2):59–61
Möglich A, Ayers RA, Moffat K (2009) Design and signaling mechanism of light-regulated histidine kinases. J Mol Biol 385(5):1433–1444
Gasser C, Taiber S, Yeh C-M, Wittig CH, Hegemann P, Ryu S et al (2014) Engineering of a red-light-activated human cAMP/cGMP-specific phosphodiesterase. Proc Natl Acad Sci U S A 111(24):8803–8808
Ryu M-H, Kang I-H, Nelson MD, Jensen TM, Lyuksyutova AI, Siltberg-Liberles J et al (2014) Engineering adenylate cyclases regulated by near-infrared window light. Proc Natl Acad Sci U S A 111(28):10167–10172
Han Y, Braatsch S, Osterloh L, Klug G (2004) A eukaryotic BLUF domain mediates light-dependent gene expression in the purple bacterium Rhodobacter sphaeroides 2.4.1. Proc Natl Acad Sci U S A 101(33):12306–12311
Strickland D, Moffat K, Sosnick TR (2008) Light-activated DNA binding in a designed allosteric protein. Proc Natl Acad Sci 105(31):10709–10714
Ohlendorf R, Vidavski RR, Eldar A, Moffat K, Möglich A (2012) From dusk till dawn: one-plasmid systems for light-regulated gene expression. J Mol Biol 416(4):534–542
Qi Y, Rao F, Luo Z, Liang Z-X (2009) A flavin cofactor-binding PAS domain regulates c-di-GMP synthesis in AxDGC2 from Acetobacter xylinum. Biochemistry 48(43):10275–10285
Barends TR, Hartmann E, Griese JJ, Beitlich T, Kirienko NV, Ryjenkov DA et al (2009) Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature 459(7249):1015–1018
Kanazawa T, Ren S, Maekawa M, Hasegawa K, Arisaka F, Hyodo M et al (2010) Biochemical and physiological characterization of a BLUF protein−EAL protein complex involved in blue light-dependent degradation of cyclic diguanylate in the purple bacterium Rhodopseudomonas palustris. Biochemistry 49(50):10647–10655
Schaap P (2013) Cyclic di-nucleotide signaling enters the eukaryote domain. IUBMB Life 65(11):897–903
Broichhagen J, Frank JA, Trauner D (2015) A roadmap to success in photopharmacology. Acc Chem Res 48(7):1947–1960
Fehrentz T, Schonberger M, Trauner D (2011) Optochemical genetics. Angew Chem Int Ed Engl 50(51):12156–12182
Luck M, Mathes T, Bruun S, Fudim R, Hagedorn R, Nguyen TMT et al (2012) A photochromic histidine kinase rhodopsin (HKR1) that is bimodally switched by ultraviolet and blue light. J Biol Chem 287(47):40083–40090
Sexton TJ, Golczak M, Palczewski K, Van Gelder RN (2012) Melanopsin is highly resistant to light and chemical bleaching in vivo. J Biol Chem 287(25):20888–20897
van Wyk M, Pielecka-Fortuna J, Lowel S, Kleinlogel S (2015) Restoring the ON switch in blind retinas: opto-mGluR6, a next-generation, cell-tailored optogenetic tool. PLoS Biol 13(5):e1002143
Beiert T, Bruegmann T, Sasse P (2014) Optogenetic activation of Gq signalling modulates pacemaker activity of cardiomyocytes. Cardiovasc Res 102(3):507–516
Avelar GM, Schumacher RI, Zaini PA, Leonard G, Richards TA, Gomes SL (2014) A rhodopsin-guanylyl cyclase gene fusion functions in visual perception in a fungus. Curr Biol 24(11):1234–1240
Scheib U, Stehfest K, Gee CE, Korschen HG, Fudim R, Oertner TG et al (2015) The rhodopsin-guanylyl cyclase of the aquatic fungus Blastocladiella emersonii enables fast optical control of cGMP signaling. Sci Signal 8(389):rs8
Gao S, Nagpal J, Schneider MW, Kozjak-Pavlovic V, Nagel G, Gottschalk A (2015) Optogenetic manipulation of cGMP in cells and animals by the tightly light-regulated guanylyl-cyclase opsin CyclOp. Nat Commun 6:8046
Ryu MH, Moskvin OV, Siltberg-Liberles J, Gomelsky M (2010) Natural and engineered photoactivated nucleotidyl cyclases for optogenetic applications. J Biol Chem 285(53):41501–41508
Balashov SP, Imasheva ES, Boichenko VA, Anton J, Wang JM, Lanyi JK (2005) Xanthorhodopsin: a proton pump with a light-harvesting carotenoid antenna. Science 309(5743):2061–2064
Wilson A, Punginelli C, Gall A, Bonetti C, Alexandre M, Routaboul J-M et al (2008) A photoactive carotenoid protein acting as light intensity sensor. Proc Natl Acad Sci U S A 105(33):12075–12080
Leverenz RL, Sutter M, Wilson A, Gupta S, Thurotte A, de Carbon CB et al (2015) A 12 angstrom carotenoid translocation in a photoswitch associated with cyanobacterial photoprotection. Science 348(6242):1463–1466
Gwizdala M, Wilson A, Kirilovsky D (2011) In vitro reconstitution of the cyanobacterial photoprotective mechanism mediated by the orange carotenoid protein in synechocystis PCC 6803. Plant Cell 23(7):2631–2643
Auldridge ME, Satyshur KA, Anstrom DM, Forest KT (2012) Structure-guided engineering enhances a phytochrome-based infrared fluorescent protein. J Biol Chem 287(10):7000–7009
Takala H, Bjorling A, Linna M, Westenhoff S, Ihalainen JA (2015) Light-induced changes in the dimerization interface of bacteriophytochromes. J Biol Chem 290(26):16383–16392
Zemelman BV, Lee GA, Ng M, Miesenböck G (2002) Selective photostimulation of genetically chARGed neurons. Neuron 33(1):15–22
Airan RD, Thompson KR, Fenno LE, Bernstein H, Deisseroth K (2009) Temporally precise in vivo control of intracellular signalling. Nature 458(7241):1025–1029
Kim JM, Hwa J, Garriga P, Reeves PJ, RajBhandary UL, Khorana HG (2005) Light-driven activation of beta 2-adrenergic receptor signaling by a chimeric rhodopsin containing the beta 2-adrenergic receptor cytoplasmic loops. Biochemistry 44(7):2284–2292
Oh E, Maejima T, Liu C, Deneris E, Herlitze S (2010) Substitution of 5-HT1A receptor signaling by a light-activated G protein-coupled receptor. J Biol Chem 285(40):30825–30836
Xiang Y, Yuan Q, Vogt N, Looger LL, Jan LY, Jan YN (2010) Light-avoidance-mediating photoreceptors tile the Drosophila larval body wall. Nature 468(7326):921–926
Liu J, Ward A, Gao J, Dong Y, Nishio N, Inada H et al (2010) C. elegans phototransduction requires a G protein-dependent cGMP pathway and a taste receptor homolog. Nat Neurosci 13(6):715–722
Ward A, Liu J, Feng Z, Xu XZ (2008) Light-sensitive neurons and channels mediate phototaxis in C. elegans. Nat Neurosci 11(8):916–922
Edwards SL, Charlie NK, Milfort MC, Brown BS, Gravlin CN, Knecht JE et al (2008) A novel molecular solution for ultraviolet light detection in Caenorhabditis elegans. PLoS Biol 6(8):e198
Bhatla N, Horvitz HR (2015) Light and hydrogen peroxide inhibit C. elegans Feeding through gustatory receptor orthologs and pharyngeal neurons. Neuron 85(4):804–818
Jost M, Fernández-Zapata J, Polanco MC, Ortiz-Guerrero JM, Chen PY-T, Kang G, Padmanabhan S, Elías-Arnanz M & Drennan CL (2015) Structural basis for gene regulation by a B12-dependent photoreceptor. Nature 526, 536–541
Muller F, Walker WH, Massey V, Brustlei M, Hemmeric P (1972) Light-absorption studies on neutral flavin radicals. Eur J Biochem 25(3):573–580
Gauden M, Yeremenko S, Laan W, van Stokkum IHM, Ihalainen JA, van Grondelle R et al (2005) Photocycle of the flavin-binding photoreceptor AppA, a bacterial transcriptional antirepressor of photosynthesis genes. Biochemistry 44(10):3653–3662
Zirak P, Penzkofer A, Lehmpfuhl C, Mathes T, Hegemann P (2007) Absorption and emission spectroscopic characterization of blue-light receptor Slr1694 from Synechocystis sp. PCC6803. J Photochem Photobiol B 86(1):22–34
Zirak P, Penzkofer A, Schiereis T, Hegemann P, Jung A, Schlichting I (2006) Photodynamics of the small BLUF protein BlrB from Rhodobacter sphaeroides. J Photochem Photobiol B 83(3):180–194
Kottke T, Heberle J, Hehn D, Dick B, Hegemann P (2003) Phot-LOV1: photocycle of a blue-light receptor domain from the green alga Chlamydomonas reinhardtii. Biophys J 84(2 Pt 1):1192–1201
Kennis JTM, Crosson S, Gauden M, van Stokkum IH, Moffat K, van Grondelle R (2003) Primary reactions of the LOV2 domain of phototropin, a plant blue-light photoreceptor. Biochemistry 42(12):3385–3392
Müller P, Bouly JP, Hitomi K, Balland V, Getzoff ED, Ritz T et al (2014) ATP binding turns plant cryptochrome into an efficient natural photoswitch. Sci Rep 4
Giovani B, Byrdin M, Ahmad M, Brettel K (2003) Light-induced electron transfer in a cryptochrome blue-light photoreceptor. Nat Struct Mol Biol 10(6):489–490
Losi A, Gensch T, van der Horst MA, Hellingwerf KJ, Braslavsky SE (2005) Hydrogen-bond network probed by time-resolved optoacoustic spectroscopy: photoactive yellow protein and the effect of E46Q and E46A mutations. Phys Chem Chem Phys 7(10):2229–2236
Lincoln CN, Fitzpatrick AE, van Thor JJ (2012) Photoisomerisation quantum yield and non-linear cross-sections with femtosecond excitation of the photoactive yellow protein. Phys Chem Chem Phys 14(45):15752–15764
Fan HY, Morgan SA, Brechun KE, Chen YY, Jaikaran ASI, Woolley GA (2011) Improving a designed photocontrolled DNA-binding protein. Biochemistry 50(7):1226–1237
Lamparter T, Esteban B, Hughes J (2001) Phytochrome Cph1 from the cyanobacterium Synechocystis PCC6803—purification, assembly, and quaternary structure. Eur J Biochem 268(17):4720–4730
Vierstra RD, Quail PH (1983) Photochemistry of 124 kilodalton Avena phytochrome in vitro. Plant Physiol 72(1):264–267
Pennacchietti F, Losi A, Xu XL, Zhao KH, Gärtner W, Viappiani C et al (2015) Photochromic conversion in a red/green cyanobacteriochrome from Synechocystis PCC6803: quantum yields in solution and photoswitching dynamics in living E. coli cells. Photochem Photobiol Sci 14(2):229–237
Popot JL, Gerchman SE, Engelman DM (1987) Refolding of bacteriorhodopsin in lipid bilayers—a thermodynamically controlled 2-stage process. J Mol Biol 198(4):655–676
Govindjee R, Balashov SP, Ebrey TG (1990) Quantum efficiency of the photochemical cycle of bacteriorhodopsin. Biophys J 58(3):597–608
Pace CN, Vajdos F, Fee L, Grimsley G, Gray T (1995) How to measure and predict the molar absorption-coefficient of a protein. Protein Sci 4(11):2411–2423
Chen D, Gibson ES, Kennedy MJ (2013) A light-triggered protein secretion system. J Cell Biol 201(4):631–640
Müller K, Engesser R, Schulz S, Steinberg T, Tomakidi P, Weber CC et al (2013) Multi-chromatic control of mammalian gene expression and signaling. Nucleic Acids Res 41(12):e124
Acknowledgments
I would like to thank John Kennis and Peter Hegemann for generous support over the last years.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer Science+Business Media New York
About this protocol
Cite this protocol
Mathes, T. (2016). Natural Resources for Optogenetic Tools. In: Kianianmomeni, A. (eds) Optogenetics. Methods in Molecular Biology, vol 1408. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-3512-3_2
Download citation
DOI: https://doi.org/10.1007/978-1-4939-3512-3_2
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-3510-9
Online ISBN: 978-1-4939-3512-3
eBook Packages: Springer Protocols