Abstract
Long interspersed nuclear element (LINE) retrotransposons make up significant parts of mammalian genomes. They alter host genomes by direct mutagenesis through integration of new transposon copies, by mobilizing non-autonomous transposons, by changes in host gene activity due to newly integrated transposons and by recombination events between different transposon copies. As a consequence, LINEs can contribute to genetic disease. Simple model systems can be useful for the study of basic molecular and cellular biology of LINE retrotransposons. Here, we describe methods for the analysis of LINE retrotransposition in the well-established model organism Saccharomyces cerevisiae. The ability to follow retrotransposition in budding yeast opens up the possibility of performing systematic screens for evolutionarily conserved interactions between LINE retrotransposons and their host cells.
Access provided by CONRICYT – Journals CONACYT. Download protocol PDF
Similar content being viewed by others
Key words
1 Introduction
Saccharomyces cerevisiae is a key model organism for molecular and genetic research. The conservation of important cellular processes between yeast and more complex eukaryotes has made yeast an attractive system for unraveling basic understanding about how cells and chromosomes replicate and divide. Some of the advantages of using yeast as a model organism are its fast division, dispersed cells, compact genome, well-defined and manipulable sexual cycle, ability to stably maintain and shuffle plasmids, and ease of modifying yeast chromosomes via homologous recombination. These traits have made yeast a premier model system for genetics and the proving ground for most new genomic technologies. Additionally, the ease of yeast chromosome manipulation has led to the construction of various useful libraries for the scientific community, such as the yeast knockout collection [1] or the yeast GFP-fusion collection [2].
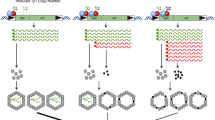
In order to utilize the technical advantages of S. cerevisiae for the study of LINE retrotransposition , we designed a system to mobilize LINE retrotransposons in budding yeast. Since the S. cerevisiae genome does not encode any known LINEs [3], we reengineered a known active LINE from Candida albicans called Zorro3 (Z3) [4]. Zorro3 is a member of the L1 clade [5, 6], and structurally similar to the currently active human and mouse L1 retrotransposons. Zorro3 retrotransposition in budding yeast is dependent on known critical amino acid residues in ORF1 and ORF2, and Zorro3 retrotransposition events cloned from the genome resemble mammalian retrotransposition events. Thus, it is likely that the mechanism of Zorro3 replication is similar to mammalian L1s, and essential host factors involved in L1 retrotransposition are conserved in budding yeast. This provides a simple tool to investigate the mechanisms by which retrotransposons and host cells interact. Additionally, the Zorro3 assay in S. cerevisiae allows experiments in a genetic background without endogenous retrotransposons of the same or closely related retrotransposon families.
Here we present a protocol to monitor Zorro3 retrotransposition in S. cerevisiae. This assay is easily adapted to different culture conditions. We also describe how to analyze and isolate retrotransposition events.
2 Materials
2.1 Yeast Strains
The described assay has been performed in the S. cerevisiae strains BY4741 ([7]; ATCC number 201388; genotype MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0) and GRF167 ([4, 8]; ATCC number 90849; genotype MATα, his3Δ200, ura3-167, GAL+) or strains derived from either of both.
2.2 Plasmids
-
1.
pSCZorro3mHIS3AI [4] contains a full-length Z3 sequence adapted to S. cerevisiae codon usage (scZ3). scZ3 is under the control of the galactose-inducible GAL1 promoter. Inserted into the 3′ untranslated region of scZ3 is an mHIS3AI cassette. This auxotrophic marker allows the selection for retrotransposition events on SC-His plates.
-
2.
pSCZorro3mHIS3AIRTmut was derived from pSCZorro3mHIS3AI by introducing the DD674,675AA mutation in the reverse transcriptase domain of ORF2. This inactive Z3 mutant was used as negative control in retrotransposition reporter assays.
2.3 Media
-
1.
YP broth: For 1 L of broth mix 10 g yeast extract, 20 g peptone, and 20 g appropriate sugar and autoclave for 15 min at 120 °C. Use dextrose for YPD broth, or galactose for YPGal.
-
2.
YP plates: Mix 1 L of YPD or YPGal broth and add 20 g bacto agar. Autoclave for 15 min at 120 °C. Let the mix cool down and pour into 10 cm petri dishes.
-
3.
Synthetic complete (SC) media broth: For 1 L mix 6.7 g yeast nitrogen base (w/o amino acids), 1.4 g appropriate dropout mix, and 20 g appropriate carbon source (dextrose or galactose) and autoclave for 15 min at 120 °C.
-
4.
SC plates: Prepare 500 mL 2× SC broth and 500 mL 4 % bacto agar. Autoclave for 15 min at 120 °C. Mix, cool down, and pour into 10 cm petri dishes (see Note 1 ).
-
5.
Dropout mix: The complete mix contains 1 g adenine, 1 g uracil, 1 g l-tryptophan, 1 g l-histidine, 1 g l-arginine, 1 g l-methionine, 3 g l-tyrosine, 4 g l-leucine, 4 g l-isoleucine, 2.5 g l-phenylalanine, 3 g l-lysine, 5 g l-glutamic acid, 5 g l-aspartic acid, 7.5 g l-valine, 10 g l-threonine, and 20 g l-serine. Use 1.4 g mix per liter of media. To make the appropriate dropout mix, omit amino acids as needed from the mix (e.g., for SC-ura omit the uracil),
-
6.
LB broth: For 1 L of broth mix 5 g yeast extract, 10 g NaCl, and 10 g bacto-tryptone. Autoclave for 15 min at 120 °C.
-
7.
LB agar plates: Prepare 1 L LB broth and add 20 g bacto agar. Autoclave for 15 min at 120 °C. Let the media cool down, add the appropriate antibiotic, and pour into 10 cm petri dishes.
2.4 Chemical Stock Solutions
-
1.
50 % PEG: Add 250 g polyethylene glycol (PEG) 3350 to 250 mL H2O. Mix and heat until the PEG is completely dissolved. Add H2O to a final volume of 500 mL. Autoclave for 15 min at 120 °C. Aliquot and store at room temperature.
-
2.
1 M lithium acetate: Add 51 g lithium acetate (Acros Organics) to 450 mL H2O. Mix until everything is dissolved. Add H2O to a final volume of 500 mL. Autoclave for 15 min at 120 °C. Aliquot and store at room temperature.
-
3.
Herring sperm DNA: Add 250 mg herring sperm DNA to 25 mL TE buffer (10 mM Tris pH 8.0, 1 mM EDTA) for a final concentration of 10 mg/mL. Mix until the DNA is completely dissolved. Aliquot and store at −20 °C.
-
4.
Acid-washed beads (Sigma).
-
5.
Lysis buffer: 100 mM NaCl, 10 mM Tris–HCl pH 7.5, 1 mM EDTA pH 8.0, 2 % Triton X-100, 1 % SDS.
Other routinely used chemicals and reagents used are obtained from local suppliers.
3 Methods
3.1 Yeast Cultivation
Yeast should be streaked from frozen stock cultures on YPD plates and incubated at 30 °C for 2–3 days before transformation. If necessary, re-streak them on another YPD plate to obtain single colonies. In general, all assays should be performed with freshly grown yeast. All strains and transformants can be stored on the workbench for several days or at 4 °C for a few weeks.
3.2 Lithium Acetate Yeast Transformation
-
1.
Inoculate 4 mL YPD with freshly grown yeast. Incubate overnight at 30 °C.
-
2.
The next morning, measure OD600. Inoculate 4 mL YPD with OD600 = 0.1/mL. Prepare one 4 mL culture for every transformation. Incubate at 30 °C until OD600 is between 0.4 and 0.5 (see Note 2 ).
-
3.
Centrifuge in a tabletop centrifuge for 5 min at 1000 × g. Discard media. Resuspend pellet in 1 mL H2O and transfer into 1.5 mL tube.
-
4.
Centrifuge for 5 min at 1000 × g. Discard H2O. Resuspend pellet in 1 mL H2O.
-
5.
Centrifuge for 5 min at 1000 × g. Discard H2O. Resuspend pellet in 1 mL 0.1 M lithium acetate.
-
6.
Centrifuge for 5 min at 1000 × g. Discard lithium acetate. Add transformation mix and mix well: 240 μL 50 % PEG, 36 μL 1 M lithium acetate, 41 μL H2O, 5 μL boiled herring sperm DNA (10 μg/mL), 2 μL plasmid DNA (0.1–0.2 μg). Incubate for 30 min at 30 °C on a rotator.
-
7.
Add 36 μL DMSO to the transformation. Mix well. Incubate for 10 min at 42 °C.
-
8.
Centrifuge for 2 min at 4000 × g. Discard supernatant. Resuspend pellet in 100 μL H2O.
-
9.
Plate 10 μL and 90 μL on selective plates (SC-ura for pZorro3mHIS3AI). Incubate at 30 °C.
-
10.
Three to four days later pick several transformants from every transformation and patch them on a new SC-ura plate. Incubate at 30 °C. Use these for retrotransposition assays.
3.3 Zorro3 Retrotransposition Assay
-
1.
Inoculate 4 mL SC-ura + galactose with freshly grown yeast transformants. Incubate at 23 °C for 3 days on a roller drum (see Notes 3 and 4 ).
-
2.
From the induced cultures, prepare at least a 10−5 dilution in H2O and plate 100 μL of this dilution on a YPD plate (see Note 5 ). The dilution should be made in serial steps. Incubate at 30 °C for 3 days. The colonies on this plate indicate the colony-forming units in the culture.
-
3.
Take a 1 mL aliquot of the induced culture and spin for 5 min at 1000 × g. Discard the media and resuspend the pellet in 150 μL H2O. Plate everything on an SC-his plate. Incubate at 30 °C for 5 days. Colonies on this plate represent retrotransposition events.
-
4.
Document the plates and count the colonies (see Note 6 ).
-
5.
Calculate the Z3 retrotransposition frequencies (see Note 7 ). The formula is (# colonies on SC-his plate)/(# colonies on YPD plate × 10 × YPD dilution).
3.4 Isolation of De Novo Zorro3 Retrotransposition Events
-
1.
Take a colony or colonies of freshly transformed with pSCzorro3mHIs3AI and patch the cells uniformly over the entire surface of a 100 mm YPGal plate. A smooth flat sterile toothpick is best for spreading the cells. Induce Z3 transcription by incubating for at least 3 days at 23 °C (see Note 8 ).
-
2.
Replica plate to SC-his and incubate for 5 days at 30 °C.
-
3.
Pick colonies from the plate to isolate genomic DNA. Inoculate 4 mL SC-his cultures with the colonies and incubate for 2 days at 30 °C. Harvest the yeast by centrifuging the cultures for 5 min at 1000 × g. Remove the media. Add 0.3 g of acid-washed beads. Resuspend the pellet in 0.2 mL lysis buffer and 0.2 mL of phenol/chloroform by vortexing for 4 min. Add 0.2 mL TE buffer and vortex briefly. Spin for 5 min at 18,000 × g and keep the top aqueous layer. Add two volumes of 100 % ethanol, mix well, and spin for 2 min at 18,000 × g. Remove the supernatant and wash the pellet with 0.75 mL 70 % ethanol. Spin again for 2 min at 18,000 × g. Remove the supernatant and air-dry the pellet for 5–10 min. Resuspend each pellet in 50 μL TE buffer. Quantify the genomic DNA by your method of choice (see Note 9 ).
-
4.
Genomic DNA digestion: Mix 0.25 μg genomic DNA, 10 U EcoRI, and 2 μL 10× EcoRI buffer in a total volume of 20 μL. Incubate overnight at 37 °C.
-
5.
Adaptor annealing: Mix 10 μL primer JH1 (0.5 mM), 10 μL primer JH122 (0.5 mM), 1 μL 5 M NaCl, and 30 μL TE buffer. Incubate for 2 min at 95 °C in a heat block. Remove the heat block and allow to gradually cool to room temperature. For primer sequences (see Note 10 ).
-
6.
Adaptor ligation: Add to the digested genomic DNA 3 μL 10× ligation buffer, 6 μL adaptor, and 1 μL (400 U) T4 DNA ligase. Ligate overnight at 4 °C.
-
7.
PCR amplification: A single reaction mix contains 2 μL ligation mix, 5 μL 10× ExTaq buffer, 4 μL dNTPs (2.5 mM each), 2.5 μL primer JH4 (5 mM), 2.5 μL JH102 (5 mM), and 0.2 μL (1 U) ExTaq DNA polymerase (Takara). Add H2O to a final volume of 50 μL. Cycling parameters: denaturation for 2 min at 94 °C, 40 cycles of 15 s at 94 °C, 15 s at 55 °C, 10 min at 72 °C, final elongation for 20 min at 72 °C (see Note 11 ).
-
8.
3′ junction cloning: Run the PCR reaction on an agarose gel and cut out the band(s). Extract the DNA with a gel extraction kit according to the manufacturer’s protocol. Elute DNA in 30 μL H2O. This product can be cloned and sequenced using any commercial or homemade TA vector (see Note 12 ).
-
9.
Once the 3′ genomic flank of the Zorro3 events are determined, the entire insertion can be PCR isolated by with primers designed to be complementary to the 3′ flank, and roughly 500 bp upstream of the 3′ flank, based on the sequenced yeast genome (see Note 13 ).
4 Notes
-
1.
Due to the low pH of SC media, SC and bacto agar are autoclaved separately to prevent hydrolysis of agar and occasional batches of “soft” plates.
-
2.
It will take 4–6 h to grow wild-type BY4741/GRF167 from OD600 = 0.1 to 0.4.
-
3.
Induction of Zorro3 transcription at temperatures higher than 23 °C results in lower retrotransposition activity.
-
4.
The retrotransposition assay should be done in at least a quadruplicate for every construct or condition. The low frequency of retrotransposition can lead to large variability.
-
5.
The end goal is to have roughly more than 100 but less than 500 colonies in the YPD plate, such that colonies can be reliably counted.
-
6.
An easy documentation and counting method is to first scan the plates. Make sure to provide a dark background that gives a good contrast to the colonies. Secondly, use an image-processing program to count the colonies. For example ImageJ is available for free and provides a handy multi-point selection tool. Always visually verify that the program is correctly marking the colonies.
-
7.
The average Z3 retrotransposition rate in the GRF167 strain is around 2 × 10−6 events/plated cells [4]. Variations are possible due to the assay conditions and the strains used.
-
8.
If the Z3 transposition frequency is much lower than in wild-type yeasts (2 × 10−6 events/plated cells) then the induction time should be extended.
-
9.
This protocol does not remove RNA, so UV absorbance will not give an accurate reading for genomic DNA. RNA can be removed with RNaseA followed by phenol/chloroform extraction.
-
10.
Adaptor primer sequences: JH1 5′-GTAATACGACTCACTATAGGGCTCCGCTTAAGGGAC-3′, JH122 5′Phos-AATTGTCCCTTAAGCGGAG-3′NH2. Different enzymes can be used to digest genomic DNA if the 5′ overhang of JH122 is altered accordingly. It is unlikely that any single enzyme/adaptor combination can be used to isolate all events.
-
11.
PCR primer sequences: JH4 5′-GTAATACGACTCACTATAGGGC-3′, JH102 5′-AGAGCTCCCGGCATCCTCTCGA-3′. JH4 hybridizes to the adaptor sequence. JH102 hybridizes to the 3′ end of scZorro3mHIS3AI. An aliquot of the PCR reaction should be loaded and separated on an agarose gel. If there are no distinct bands then a second PCR can be performed with either the same primers or nested primers and 2 μL of the first PCR as template.
-
12.
Sometimes the polyA tail will be too long to sequence through. In this case, sequence adjacent to the polyA tail can be obtained by sequencing with anchored polyA primers (A)19T, (A)19C, and (A)19G.
-
13.
Note that the majority of retrotransposition events produced in the Zorro3 assay result in episomal circles [9]. These circles tend to be unstable under nonselective conditions. To enrich for chromosomally integrated events, the SC-ura + galactose plates can be replica plated to YPD plates and incubated for 48 h at 30 °C prior to replica plating to SC-his plates. Chromosomally integrated events will tend to form the larger colonies on the SC-his plates.
References
Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, Bakkoury El M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M’Rabet N, Menard P, Mittmann M, Pai C, Rebischung C, Revuelta JL, Riles L, Roberts CJ, Ross-MacDonald P, Scherens B, Snyder M, Sookhai-Mahadeo S, Storms RK, Véronneau S, Voet M, Volckaert G, Ward TR, Wysocki R, Yen GS, Yu K, Zimmermann K, Philippsen P, Johnston M, Davis RW (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901–906
Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425:686–691
Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG (1996) Life with 6000 genes. Science 274:546–563
Dong C, Poulter RT, Han JS (2009) LINE-like retrotransposition in Saccharomyces cerevisiae. Genetics 181:301–311
Goodwin TJ, Ormandy JE, Poulter RT (2001) L1-like non-LTR retrotransposons in the yeast Candida albicans. Curr Genet 39:83–91
Goodwin TJ, Busby JN, Poulter RT (2007) A yeast model for target-primed (non-LTR) retrotransposition. BMC Genomics 8:263
Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14(2):115–132
Boeke JD, Garfinkel DJ, Styles CA, Fink GR (1985) Ty elements transpose through an RNA intermediate. Cell 40:491–500
Han JS, Shao S (2012) Circular retrotransposition products generated by a LINE retrotransposon. Nucleic Acids Res 40:10866–10877
Acknowledgements
This work was supported by NIH grant GM090192 to J.S.H.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer Science+Business Media New York
About this protocol
Cite this protocol
Horn, A.V., Han, J.S. (2016). LINE Retrotransposition Assays in Saccharomyces cerevisiae . In: Garcia-Pérez, J. (eds) Transposons and Retrotransposons. Methods in Molecular Biology, vol 1400. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-3372-3_9
Download citation
DOI: https://doi.org/10.1007/978-1-4939-3372-3_9
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-3370-9
Online ISBN: 978-1-4939-3372-3
eBook Packages: Springer Protocols