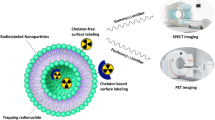
Abstract
Ever since the discovery of RNA interference (RNAi), therapeutic delivery of siRNA has attracted a lot of interest. However, due to the nature and structure of siRNA, a carrier is needed for any mode of systemic treatment. Furthermore, specific imaging techniques are required to trace where the deposition of the siRNA occurs throughout the body after treatment. Tracking in vivo siRNA biodistribution allows understanding and interpreting therapeutics effects and side effects. A great advantage of noninvasive imaging techniques such as SPECT imaging is that several time points can be assessed in the same subject. Thus, the time course of biodistribution or metabolic processes can be followed. Therefore, we have described an approach to modify siRNA with a DTPA (Diethylene Triamine Pentaacetic Acid) chelator in order to utilize an indium labeled siRNA for SPECT imaging. Here, we explain the details of the labeling and purification procedures.
Access provided by CONRICYT – Journals CONACYT. Download protocol PDF
Similar content being viewed by others
Key words
1 Introduction
Since the Nobel Prize in physiology was awarded in 2006 to Andrew Fire and Greg Mello for their work in RNA interference (RNAi), there has been an increase in the development of RNAi as a therapeutic tool to transiently knock down specific proteins. Unfortunately when it is delivered in vivo, naked small interfering RNA (siRNA ) is taken up very poorly into the cell due to its molecular makeup [1–4]. To improve its uptake and specificity, siRNA can for example be packaged inside nanoparticles. Nanoparticle delivery offers several benefits such as increased stability, longer circulation time, capability to package multiple payloads, and specific targeting to tumor sites [5, 6].
The most common routes of nanoparticle administration for therapeutic use are intravenous, transdermal, pulmonary administration, and intraocular [7–9]. For cancer therapy, intravenous delivery of the nanoparticles is most ideal due to the ability of the nanoparticles to inherently preferentially reach the tumors directly from the bloodstream due to the enhanced permeation and retention (EPR) effect [10]. However, when nanoparticles are administered intravenously , several obstacles need to be overcome. Among those obstacles are the circulation profile and deposition of the nanoparticles. Both of those are key components to the success of any treatment.
In order to assess the efficacy of the nanoparticle deposition in the organ or tissue of interest, single photon emission computed tomography (SPECT ) imaging can be used. SPECT imaging is often employed to detect radioactive species within the body [11]. More specifically, for siRNA delivery, this can be utilized to trace where radioactive siRNA travels throughout the bloodstream and where specifically it deposits inside the body. This approach provides useful information about the obstacles that are needed to be overcome for intravenous delivery. Consequently, SPECT imaging can illustrate whether the siRNA loaded nanoparticles are degraded in circulation before reaching the tumor site, as well as its biodistribution [12, 13]. Due to the need to overcome these hurdles for a successful treatment, siRNA imaging techniques are needed.
This chapter outlines the technique to label siRNA with a DTPA chelator. Once the siRNA is sufficiently labeled with DTPA, it can be reacted with indium and annealed in order to become radioactive. Once the indium has been chelated to the DTPA, the siRNA can then be packaged inside a nanoparticle or other nanocarriers and imaged with a SPECT scanner.
2 Materials
2.1 siRNA Formulation
Due to their increased stability, high activity, and ability to be covalently modified, 2′-O-methylated 25/27mer DsiRNA targeting EGFP [14] from Integrated DNA Technologies (Leuven, Belgium) was used and is recommended for use. For coupling of DTPA , amine-labeled siRNA is recommended. Here, we used a duplex with an amino-hexyl modification at the 5-prime of the antisense strand.
-
1.
siEGFP: sense: 5′-pACCCUGAAGUUCAUCUGCACCACdCdG, antisense: 3′-mAmCmUGmGGmACmUUmCAmAGmUAmGAmCGUGGUGGC-C6H12NH2
2.2 Covalent Modification of siRNA with pBn-SCN-Bn-DTPA
-
1.
siEGFP—described above.
-
2.
pBn-SCN-Bn-DTPA (Macrocyclics).
-
3.
0.1 M NaHCO3 in DEPC water—filtered through a 0.22 μm filter before use.
-
4.
2M NaOAc in DEPC water—filtered through a 0.22 μm filter before use.
-
5.
Dried DMSO (about 3 mL).
-
6.
0.22 μm filter.
-
7.
2 mL centrifuge tube.
-
8.
Metal spatula wrapped in parafilm—Used to weigh out pBn-SCN-Bn-DTPA (see Note 1 ).
-
9.
Aluminum foil.
-
10.
Vortex.
2.3 Precipitation of the siRNA–DTPA Complex
-
1.
2M NaOAc in RNase free water—filtered through a 0.22 μm filter before use.
-
2.
Absolute ethanol—filtered through a 0.22 μm filter before use.
-
3.
15 mL conical tube.
2.4 Isolation of the siRNA–DTPA Complex
-
1.
Ultracentrifuge (see Note 6 ).
-
2.
Lysis Buffer from Absolutely RNA miRNA Kit (Agilent).
-
3.
2M NaOAc in RNase free water—filtered through a 0.22 μm filter before use.
-
4.
Absolute ethanol—filtered through a 0.22 μm filter before use.
-
5.
5 RNeasy Midi Kit (10) columns (Qiagen).
2.5 siRNA–DTPA Purification
-
1.
Centrifuge.
-
2.
Low salt buffer from the Absolutely RNA miRNA Kit (Agilent).
-
3.
RNase free water.
-
4.
Sterile 2 mL collection centrifuge tubes.
2.6 siRNA Concentration Measurement
-
1.
Nanodrop.
-
2.
RNase free water.
-
3.
0.5 mL tubes.
-
4.
Parafilm.
-
5.
Dry heat bath set to 94 °C.
-
6.
Timer.
2.7 DTPA Concentration Measurement
-
1.
Stock solution of the yttrium(III)–arsenazo III complex containing 5 μM arsenazo(III) (Chem-Impex INT’L INC.), and 1.6 μM yttrium(III) chloride (Acros Organics) in a 0.15 M NaOAc buffer at pH 4.
-
2.
Stock solution of 0.123 mM DTPA dissolved in DI-H2O with 3 molar equivalents of NaOH (see Note 2 ).
-
3.
UV–Vis spectrophotometer.
-
4.
UV–Vis disposable cuvette.
2.8 Indium Labeling
-
1.
Radioactive Indium (III) chloride (Covidien).
-
2.
GE Healthcare Disposable PD-10 Desalting Columns.
-
3.
RNase free water.
-
4.
Scintillation vials (make and model to fit gamma counter).
-
5.
Gamma counter (e.g., Packard 5005).
-
6.
Nanodrop.
2.9 Animal Imaging
-
1.
Mice, e.g., 6-week-old balb/c mice.
-
2.
Sterile Insulin Syringes.
-
3.
Sterile siRNA formulation, e.g., nanoparticles.
-
4.
Mouse anesthesia.
-
5.
SPECT imaging device and mouse cradle, e.g., Siemens E.CAM with custom-made collimator.
3 Methods
3.1 React siRNA with pBn-SCN-DTPA
-
1.
Weigh out 5.11 mg of Double Stranded siRNA in a 2 mL centrifuge tube and dissolve it in 100 μL of RNase free water (see Note 3 ).
-
2.
To the 2 mL tube, add 100 μL of filtered 0.1 M NaHCO3.
-
3.
Next, weigh out 9.76 mg of pBn-SCN-Bn-DTPA and dissolve it in 540 μL of dry DMSO (see Notes 1 and 4 ).
-
4.
Add the 540 μL of the DTPA to siRNA mixture. The new total volume should be 740 μL (see Note 5 ).
-
5.
Wrap the solution in tin foil, vortex thoroughly, and incubate for 6 h. Agitate the solution every 30 min.
3.2 Precipitation of the siRNA–DTPA Complex
-
1.
Add 74 μL of filtered 2 M NaOAc to the mixture (10 % of the total amount of mixture).
-
2.
Transfer the mixture to a 15 mL conical tube.
-
3.
Add filtered absolute ethanol so that the final concentration is 80 % v/v.
-
4.
Freeze solution overnight at −80 °C.
3.3 Isolation of the siRNA–DTPA Complex
-
1.
Centrifuge the sample for 30 min at 12,000 × g in an ultracentrifuge (see Note 6 ).
-
2.
Discard the supernatant.
-
3.
Add 2.5 mL of Lysis Buffer from “Absolutely RNA miRNA Kit” (Agilent).
-
4.
Vortex the solution.
-
5.
Add 250 μL of filtered 2M NaOAc.
-
6.
Add 7.25 mL of filtered absolute ethanol for a total of 10 mL (see Note 7 ).
-
7.
Vortex the solution and equally distribute the 10 mL onto 5 RNeasy Midi Kit (10) Qiagen Columns (see Note 8 ).
3.4 siRNA–DTPA Purification
-
1.
Centrifuge the columns at 4500 × g for 5 min, discard the flow through.
-
2.
To each column, add 200 μL of the low salt buffer from the Absolutely RNA miRNA Kit (Agilent).
-
3.
Centrifuge the solution at 4500 × g for 2 min, discard the flow through.
-
4.
Repeat steps 2 and 3.
-
5.
To dry the column, spin them down at 5000 × g for 5 min.
-
6.
Transfer the columns to a new collection tube and add 200 μL of 60 °C hot RNase free water.
-
7.
Centrifuge the solution at 5000 × g for 5 min to collect the purified siRNA –DTPA .
-
8.
Add 100 μL of 60 °C hot RNase free water and centrifuge at 5000 × g for 5 min.
-
9.
Combine the flow through from all of the columns into one sterile 2 mL tube.
3.5 siRNA Concentration Measurement
-
1.
Measure the siRNA concentration on a nanodrop (Thermo Scientific—Nanodrop 2000c). Use RNase free water as your blank.
-
2.
In a sterile environment, dilute the siRNA to a desired concentration and aliquot into 0.5 mL sterile tubes (see Note 9 ).
-
3.
Filter the siRNA –DTPA solution to make it sterile.
-
4.
Parafilm each tube and anneal the siRNA at 94 °C for exactly 2 min.
-
5.
Let the samples cool down to room temperature.
-
6.
Freeze the samples and keep frozen until needed.
3.6 DTPA Concentration Measurement
-
1.
Using a UV–Vis spectrophotometer (Cary—50 Bio), you will create the standard curve for DTPA concentrations.
-
2.
Pipette 3 mL of the Y(III)–arsenazo III complex stock solution into a cuvette and read (652 nM) this as the blank.
-
3.
Add 5 μL of the stock DTPA solution to the cuvette, gently mix, and read the solution again (see Note 10 ).
-
4.
Add another 5 μL of the stock DTPA solution, read, and repeat.
-
5.
Do this until you have generated enough points for your standard curve.
-
6.
Each new data point will have an additional 5 μL added into the cuvette.
-
7.
Once all the standards have been made and read on the spectrophotometer, discard the solution inside the cuvette and put 3 mL of fresh Y(III)–arsenazo III complex stock solution into the cuvette (see Note 11 ).
-
8.
Add 5 μL of your siRNA –DTPA sample into the 3 mL and take the measurement.
-
9.
Plot the standard curve for the DTPA concentrations versus absorbance and insert a linear line of best fit (Fig. 1).
Fig. 1 -
10.
Using the equation yielded from the line of best fit, plug the absorbance value obtained from your sample measurement into the Y-value of the equation in order to solve for X.
-
11.
The X-value obtained will be the concentration of DTPA in your sample.
-
12.
Now that the DTPA and siRNA concentrations have been found for the siRNA–DTPA mixture, figure out the molar amounts of the siRNA and DTPA within your sample. From here, you can determine the molar equivalency of the siRNA and DTPA (see Note 12 ).
3.7 Indium Labeling and Purification
-
1.
React radioactive 111InCl3 with siRNA . In the example shown below, 116.9 MBq 111InCl3 were reacted with 15 nmol siRNA. Incubate for 30 min at room termperature.
-
2.
Equilibrate a PD-10 column with RNase free water by washing it with 25 ml.
-
3.
Prepare 24 scintillation vials in a rack and label them from 1 to 24.
-
4.
Place vial 1 underneath the PD-10 column and start adding the siRNA –Indium mixture to the column slowly.
-
5.
Collect 13 drops in the first vial as fraction 1 and then move on to the next vial. Collect 13 drops per fraction. Once the complete volume of the siRNA –Indium mixture is applied to the column, add RNase free water. Collect 24 fractions.
-
6.
Close the scintillation vials and measure the counts per minute (CPM) in every vial using a gamma counter.
-
7.
Plot the CPM versus the fraction number (Fig. 2).
-
8.
Determine the siRNA concentration in the peak fraction using a Nanodrop (Note 13 ).
3.8 Animal Imaging
-
1.
Prepare the siRNA formulation to be administered, e.g., nanoparticles. In the example below, micelles of polyethyleneimine-graft-polycaprolactone-block-poly(ethylene glycol) were prepared with 2 nmol siRNA per animal which was equivalent to approximately 3 MBq per animal.
-
2.
Anesthetize the animals and administer the siRNA formulation. In the example below, injection to the tail vein was chosen.
-
3.
Place the animals, one after the other, in the cradle and start the 360° imaging program (Fig. 3) (Note 14 ).
Fig. 3 SPECT images of a 6-week-old balb/c mouse i.v. injected with 2.9 MBq 111Indium-labeled siRNA formulated as micelles 2 h after injection. The formulation was obtained with a polymer carrying a short, 500 Da, PEG chain which is the reason for the accumulation of the micelles in the lung. Deposition in the liver and excretion through the kidneys into the bladder can be observed as well
4 Notes
-
1.
Wrap the metal spatula in parafilm so the DTPA does not complex to the metal ions from the spatula.
-
2.
You should first dissolve the DTPA in DMSO before diluting in the DI-H2O with NaOH. Make sure the DMSO is at least diluted out by a factor of 1:100.
-
3.
Our siRNA had an MW of 17950.36 g/mol. Therefore, we use 0.285 μmol of siRNA.
-
4.
Total pBn-SCN-DTPA (MW = 649.9 g/mol) is 15.02 μmol.
-
5.
The solution turned cloudy upon the addition of the DTPA to the siRNA solution.
-
6.
You should get a nice visible white pellet at the bottom of the 15 mL conical tube. A regular centrifuge that reaches a speed of 12,000 × g may as well be used.
-
7.
Upon addition of the ethanol, the solution should turn slightly cloudy again.
-
8.
You should put roughly 2 mL into each column due to the fact that each column can only retain 1 mg RNA and a limited volume. If you add too much, you may lose some during the purification steps.
-
9.
To make calculations easier in the future, dilute the siRNA to either 100 mM or 50 mM. Aliquot the samples into small portions to prevent several freeze–thaw cycles.
-
10.
Mix the samples by gently pipetting up and down within the cuvette. Be careful not to create any bubbles.
-
11.
Make sure you rinse out the cuvette very well. When you read the fresh 3 mL of the complex solution, verify that the values are in line with the previous measurements.
-
12.
Since each siRNA strand has only 1 amine group for DTPA to complex to, if performed correctly, your ratio should be approximately a 1:1 molar equivalence of DTPA and siRNA. If the ratio of DTPA per siRNA is higher than 1:1, residual free DTPA was not removed during the purification.
-
13.
It may be necessary to combine 2 or more peak fractions based on the CPM values and RNA concentrations. If free DTPA is present in the siRNA solution when it is radiolabeled, a second small peak will appear around fraction 12, and free Indium appears around fraction 20.
-
14.
The imaging procedure can be repeated at any given time. The half-life of 111Indium is 2.6 days, and a significant amount of siRNA is typically excreted renally or even hepatically. Therefore, imaging at time points later than 48 h can become challenging.
References
Dickerson EB, Blackburn WH, Smith MH, Kapa LB, Lyon LA, McDonald JF (2010) Chemosensitization of cancer cells by siRNA using targeted nanogel delivery. BMC Cancer 10:10
Huang YH, Bao Y, Peng W, Goldberg M, Love K, Bumcrot DA, Cole G, Langer R, Anderson DG, Sawicki JA (2009) Claudin-3 gene silencing with siRNA suppresses ovarian tumor growth and metastasis. Proc Natl Acad Sci U S A 106(9):3426–3430
Shahzad MM, Lu C, Lee JW, Stone RL, Mitra R, Mangala LS, Lu Y, Baggerly KA, Danes CG, Nick AM, Halder J, Kim HS, Vivas-Mejia P, Landen CN, Lopez-Berestein G, Coleman RL, Sood AK (2009) Dual targeting of EphA2 and FAK in ovarian carcinoma. Cancer Biol Ther 8(11):1027–1034
Goldberg MS, Xing D, Ren Y, Orsulic S, Bhatia SN, Sharp PA (2011) Nanoparticle-mediated delivery of siRNA targeting Parp1 extends survival of mice bearing tumors derived from Brca1-deficient ovarian cancer cells. Proc Natl Acad Sci U S A 108(2):745–750
Vlahov I, Leamon CP (2012) Engineering folate-drug conjugates to target cancer: from chemistry to clinic. Bioconjug Chem 23:1357–1369
Akhtar S (2006) Non-viral cancer gene therapy: beyond delivery. Gene Ther 13:739–740
Watts JK, Corey DR (2010) Clinical status of duplex RNA. Bioorg Med Chem Lett 20:3203–3207
Durcan N, Murphy C, Cryan SA (2008) Inhalable siRNA: potential as a therapeutic agent in the lungs. Mol Pharm 5:559–566
Davies LA et al (2007) Identification of transfected cell types following non-viral gene transfer to the murine lung. J Gene Med 9:184–196
Bertrand N, Wu J, Xu X, Kamaly N, Farokhzad O (2014) Cancer nanotechnology: the impact of passive and active targeting in the era of modern cancer biology. Adv Drug Deliv Rev 66:2–25
Renette T, Librizzi D, Endres T, Merkel O, Broichsitter MB, Bege N, Petersen H, Curdy C, Kissel T (2012) Poly(ethylene carbonate) nanoparticles as carrier system for chemotherapy showing prolonged in vivo circulation and anti-tumor efficacy. Macromol Biosci 12:970–978
Pippin CG, Parker TA, McMurry TJ, Brechbiel MW (1992) Spectrophotometric method for the determination of a bifunctional DTPA ligand in DTPA-monoclonal antibody conjugates. Bioconjug Chem 3:342–345
Merkel OM, Librizzi D, Pfestroff A, Schurrat T, Behe M, Kissel T (2009) In vivo SPECT and real-time gamma camera imaging of biodistribution and pharmacokinetics of siRNA delivery. Bioconjug Chem 20:174–182
Rose SD et al (2005) Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Res 33:4140–4156
Acknowledgements
This work was supported by the NanoIncubator grant to Olivia Merkel as well as the Ruth L. Kirschstein National Research Award T32-CA009531 fellowship to Steven Jones.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer Science+Business Media New York
About this protocol
Cite this protocol
Jones, S., Merkel, O. (2016). Indium-Labeling of siRNA for Small Animal SPECT Imaging. In: Medarova, Z. (eds) RNA Imaging. Methods in Molecular Biology, vol 1372. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-3148-4_6
Download citation
DOI: https://doi.org/10.1007/978-1-4939-3148-4_6
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-3147-7
Online ISBN: 978-1-4939-3148-4
eBook Packages: Springer Protocols