Abstract
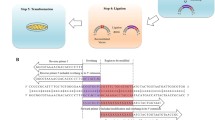
Inverse PCR is a powerful tool for the rapid introduction of desired mutations at desired positions in a circular double-stranded DNA sequence. In this technique, custom-designed mutant primers oriented in the inverse direction are used to amplify the entire circular template with incorporation of the required mutation(s). By careful primer design, it can be used to perform such diverse modifications as the introduction of point or multiple mutations, the insertion of new sequences, and even sequence deletions. Three primer formats are commonly used, nonoverlapping, partially overlapping, and fully overlapping primers, and here we describe the use of nonoverlapping primers for introduction of a point mutation. Use of such a primer setup in the PCR, with one of the primers containing the desired mismatch mutation, results in the amplification of a linear, double-stranded, mutated product. Methylated template DNA is removed from the non-methylated PCR product by DpnI digestion, and the PCR product is then phosphorylated by polynucleotide kinase treatment before being recircularized by ligation and transformed to E. coli. This relatively simple site-directed mutagenesis procedure is of major importance in biology and biotechnology where it is commonly employed for the study and engineering of DNA, RNA, and proteins.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Burdick JP, Basi RS, Burns KS, Weers PMM (1865) The role of C-terminal ionic residues in self-association of apolipoprotein A-I. Biochim Biophys Acta Biomembr 2023:184098. https://doi.org/10.1016/j.bbamem.2022.184098
Dahhas MA, Alsenaidy MA (2022) Role of site-directed mutagenesis and adjuvants in the stability and potency of anthrax protective antigen. Saudi Pharm J 30:595–604. https://doi.org/10.1016/j.jsps.2022.02.011
Wang L, Gu J, Zhao W, Wang M, Ng KR, Lyu X, Yang R (2022) Reshaping the binding pocket of cellobiose 2-epimerase for improved substrate affinity and isomerization activity for enabling green synthesis of lactulose. J Agric Food Chem 70:15879–15893. https://doi.org/10.1021/acs.jafc.2c06980
Chen K, Zhang M, Gao B, Hasan A, Li J, Bao Y, Fan J, Yu R, Yi Y, Ågren H, Wang Z, Liu H, Ye M, Qiao X (2022) Characterization and protein engineering of glycosyltransferases for the biosynthesis of diverse hepatoprotective cycloartane-type saponins in Astragalus membranaceus. Plant Biotechnol J 21:698. https://doi.org/10.1111/pbi.13983
Sanguinetti M, Silva Santos LH, Dourron J, Alamón C, Idiarte J, Amillis S, Pantano S, Ramón A (2022) Substrate recognition properties from an intermediate structural state of the UreA transporter. Int J Mol Sci 23:16039. https://doi.org/10.3390/ijms232416039
Wang K-D, Dughbaj MA, Nguyen TTV, Nguyen TQY, Oza S, Valdez K, Anda P, Waltz J, Sacco MA (2023) Systematic mutagenesis of Polerovirus protein P0 reveals distinct and overlapping amino acid functions in Nicotiana glutinosa. Virology 578:24–34. https://doi.org/10.1016/j.virol.2022.11.005
Collins T, De Vos D, Hoyoux A, Savvides SN, Gerday C, Van Beeumen J, Feller G (2005) Study of the active site residues of a glycoside hydrolase family 8 xylanase. J Mol Biol 354:425–435. https://doi.org/10.1016/j.jmb.2005.09.064
Hutchison CA, Phillips S, Edgell MH, Gillam S, Jahnke P, Smith M (1978) Mutagenesis at a specific position in a DNA sequence. J Biol Chem 253:6551–6560
Edgell MH, Hutchison CA 3rd, Sclair M (1972) Specific endonuclease R fragments of bacteriophage phiX174 deoxyribonucleic acid. J Virol 9:574–582
Hutchison CA 3rd, Edgell MH (1971) Genetic assay for small fragments of bacteriophage phi X174 deoxyribonucleic acid. J Virol 8:181–189
Reeves AR (2016) In Vitro Mutagenesis. Methods Protocol:1–796
Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59
Das N, Ghosh Dhar D, Dhar P (2022) Editing the genome of common cereals (rice and wheat): techniques, applications, and industrial aspects. Mol Biol Rep:1–9. https://doi.org/10.1007/S11033-022-07664-Y/TABLES/2
Forner J, Kleinschmidt D, Meyer EH, Fischer A, Morbitzer R, Lahaye T, Schöttler MA, Bock R (2022) Targeted introduction of heritable point mutations into the plant mitochondrial genome. Nat Plants 8:245–256. https://doi.org/10.1038/s41477-022-01108-y
Guo J, Zeng L, Chen H, Ma C, Tu J, Shen J, Wen J, Fu T, Yi B (2022) CRISPR/Cas9-mediated targeted mutagenesis of BnaCOL9 advances the flowering time of Brassica napus L. Int J Mol Sci 23:14944. https://doi.org/10.3390/ijms232314944
Higuchi R, Krummel B, Saiki RK (1988) A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res 16:7351–7367
Triglia T, Peterson MG, Kemp DJ (1988) A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res 16:8186
Ochman H, Gerber AS, Hartl DL (1988) Genetic applications of an inverse polymerase chain reaction. Genetics 120:621–623
Hemsley A, Arnheim N, Toney MD, Cortopassi G, Galas DJ (1989) A simple method for site-directed mutagenesis using the polymerase chain reaction. Nucleic Acids Res 17:6545–6551
Qi D, Scholthof KB (2008) A one-step PCR-based method for rapid and efficient site-directed fragment deletion, insertion, and substitution mutagenesis. J Virol Methods 149:85–90. https://doi.org/10.1016/j.jviromet.2008.01.002
Zheng L, Baumann U, Reymond JL (2004) An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res 32:e115. https://doi.org/10.1093/nar/gnh110
Liu H, Naismith JH (2008) An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol 8:91. https://doi.org/10.1186/1472-6750-8-91
Xia Y, Chu W, Qi Q, Xun L (2015) New insights into the QuikChange process guide the use of Phusion DNA polymerase for site-directed mutagenesis. Nucleic Acids Res 43:e12. https://doi.org/10.1093/nar/gku1189
Silva D, Santos G, Barroca M, Collins T (2017) Inverse PCR for point mutation introduction. In: Domingues L (ed) PCR. Methods and protocols, Springer protocols. Methods in molecular biology, 1st edn. Springer New York, New York, pp 87–100. https://doi.org/10.1007/978-1-4939-7060-5_5
Acknowledgments
The European Regional Development Fund (ERDF) is thanked for funding in the scope of Programa Operacional Regional do Norte (NORTE 2020) through the project ATLANTIDA (NORTE-01-0145-FEDER-000040). The FCT (Fundação para a Ciência e a Tecnologia) is thanked for funding through the “Contrato-Programa” UIDB/04050/2020.
All the technical staff at the CBMA are thanked for their skillful technical assistance.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Silva, D., Santos, G., Barroca, M., Costa, D., Collins, T. (2023). Inverse PCR for Site-Directed Mutagenesis. In: Domingues, L. (eds) PCR. Methods in Molecular Biology, vol 2967. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-3358-8_18
Download citation
DOI: https://doi.org/10.1007/978-1-0716-3358-8_18
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-3357-1
Online ISBN: 978-1-0716-3358-8
eBook Packages: Springer Protocols