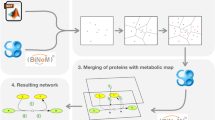
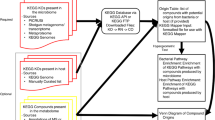
Abstract
Alteration of the status of the metabolic enzymes could be a probable way to regulate metabolic reprogramming, which is a critical cellular adaptation mechanism especially for cancer cells. Coordination among biological pathways, such as gene-regulatory, signaling, and metabolic pathways is crucial for regulating metabolic adaptation. Also, incorporation of resident microbial metabolic potential in human body can influence the interplay between the microbiome and the systemic or tissue metabolic environments. Systemic framework for model-based integration of multi-omics data can ultimately improve our understanding of metabolic reprogramming at holistic level. However, the interconnectivity and novel meta-pathway regulatory mechanisms are relatively lesser explored and understood. Hence, we propose a computational protocol that utilizes multi-omics data to identify probable cross-pathway regulatory and protein-protein interaction (PPI) links connecting signaling proteins or transcription factors or miRNAs to metabolic enzymes and their metabolites using network analysis and mathematical modeling. These cross-pathway links were shown to play important roles in metabolic reprogramming in cancer scenarios.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Warburg O (1925) The metabolism of carcinoma cells. J Cancer Res 9:148–163
Warburg O (1956) On the origin of cancer cells. Science 123:309–314
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144:646–674
Patrick S, Ward CBT (2013) Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell 21:297–308
Ward CBT, Patrick S, Thompson CB (2012) Signaling in control of cell growth and metabolism. Cold Spring Harb Perspect Biol 4:a006783
Mo Y, Wang Y, Zhang L et al (2019) The role of Wnt signaling pathway in tumor metabolic reprogramming. J Cancer 10:3789–3797
Papa S, Choy PM, Bubici C (2019) The ERK and JNK pathways in the regulation of metabolic reprogramming. Oncogene 38:2223–2240
Martin-Martin N, Carracedo A, Torrano V (2017) Metabolism and transcription in cancer: merging two classic tales. Front Cell Dev Bio 5:119
Dong Y, Tu R, Liu H et al (2020) Regulation of cancer cell metabolism: oncogenic MYC in the driver’s seat. Sig Transduct Target Ther 5:124
Machida K (2018) Pluripotency transcription factors and metabolic reprogramming of mitochondria in tumor-initiating stem-like cells. Antioxid Redox Signal 28:1080–1089
Rottiers V, Naar AM (2012) MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol 13:239–250
Chen B, Li H, Zeng X et al (2012) Roles of microRNA on cancer cell metabolism. J Transl Med 10:228
Singh PK, Mehla K, Hollingsworth MA et al (2011) Regulation of aerobic glycolysis by microRNAs in cancer. Mol Cell Pharmacol 3:125–134
Fung TC, Olson CA, Hsiao EY (2017) Interactions between the microbiota, immune and nervous systems in health and disease. Nat Neurosci 20:145–155
Zheng D, Liwinski T, Elinav E (2020) Interaction between microbiota and immunity in health and disease. Cell Res 30:492–506
Gopalakrishnan V, Helmink BA, Spencer CN et al (2018) The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell 33:570–580
Helmink BA, Khan MAW, Hermann A et al (2020) The microbiome, cancer, and cancer therapy. Nat Med 25:377–388
Xavier JB, Young VB, Skufka J et al (2020) The cancer microbiome: distinguishing direct and indirect effects requires a systemic view. Trends Cancer 6:192–204
Szklarczyk D, Gable AL, Lyon D et al (2019) STRING v11 : protein –protein association networks with increased coverage , supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47:D607–D613
Kanehisa M, Sato Y, Kawashima M et al (2016) KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44:D457–D462
Fabregat A, Sidiropoulos K, Garapati P et al (2016) The Reactome pathway knowledgebase. Nucleic Acids Res 44:D481–D487
Croft D, Mundo AF, Haw R et al (2014) The Reactome pathway knowledgebase. Nucleic Acids Res 42:D472–D477
Fazekas D, Koltai M, Türei D et al (2013) SignaLink 2 – a signaling pathway resource with multi-layered regulatory networks. BMC Syst Biol 7:7
Kandasamy K, Mohan SS, Raju et al (2010) NetPath: a public resource of curated signal transduction pathways. Genome Biol 11:R3
Bovolenta LA, Acencio ML, Lemke N (2012) HTRIdb: an open-access database for experimentally verified human transcriptional regulation interactions. BMC Genomics 13:405
Han H, Cho JW, Lee S et al (2018) TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res 46:D380–D386
Huang HY, Lin YCD, Li J et al (2020) miRTarBase 2020: updates to the experimentally validated microRNA–target interaction database. Nucleic Acids Res 48:D148–D154
Karagkouni D, Paraskevopoulou MD, Chatzopoulos S et al (2018) DIANA-TarBase v8:a decade-long collection of experimentally supported miRNA-gene interaction. Nucleic Acids Res 46:D239–D245
Mandloi S, Chakrabarti S (2015) PALM-IST: pathway assembly from literature mining--an information search tool. Sci Rep 5:10021
Hoffmann R, Valencia A (2004) A gene network for navigating the literature. Nat Genet 36:664
Shah PK, Jensen LJ, Boue S, Bork P (2005) Extraction of transcript diversity from scientific literature. PLoS Comput Biol 1(1):e10
Horn F, Lau AL, Cohen FE (2004) Automated extraction of mutation data from the literature: application of MuteXt to G protein-coupled receptors and nuclear hormone receptors. Bioinformatics 20:557–568
Hu ZZ, Narayanaswamy M, Ravikumar KE, Vijay-Shanker K, Wu CH (2005) Literature mining and database annotation of protein phosphorylation using a rule-based system. Bioinformatics 21:2759–2765
https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga
Zhang J, Baran J, Cros A et al (2011) International Cancer Genome Consortium Data Portal-a one-stop shop for cancer genomics data. Database 2011:bar026
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11:R106
Hardcastle TJ (2021) baySeq: Empirical Bayesian analysis of patterns of differential expression in count data. R package version 2.26.0
Leng N, Kendziorski C (2021) EBSeq: an R package for gene and isoform differential expression analysis of RNA-seq data. R package version 1.32.0
Barrett T, Wilhite SE, Ledoux P et al (2013) NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res 41:D991–D995
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Ser B 57:289–300
Huson DH, Auch AF, Qi J et al (2007) MEGAN analysis of metagenomic data. Genome Res 17:377–386
Schloss PD, Westcott SL, Ryabin T et al (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Caporaso JG, Kuczynski J, Stombaugh J et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461
Rognes T, Flouri T, Nichols B et al (2016) Vsearch: a versatile open source tool for metagenomics. PeerJ 4:e2584
Albanese D, Fontana P, De Filippo C et al (2015) MICCA: a complete and accurate software for taxonomic profiling of metagenomic data. Sci Rep 5:1–7
Bolyen E, Rideout JR, Dillon MR et al (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857
Callahan BJ, McMurdie PJ, Rosen MJ et al (2016) DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583
DeSantis TZ, Hugenholtz P, Larsen N et al (2006) Greengenes, a Chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072
Quast C, Pruesse E, Yilmaz P et al (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596
Cole JR, Wang Q, Fish JA et al (2014) Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42:D633–D642
Pedregosa F, Varoquaux G, Gramfort A et al (2011) Scikit-learn: machine learning in Python. J Machine Learn Res 12:2825–2830
Robeson MS, O’Rourke DR, Kaehler BD et al (2021) RESCRIPt: reproducible sequence taxonomy reference database management for the masses. PLoS Comput Biol e1009581
Langille MGI, Zaneveld J, Caporaso JG et al (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814
Douglas GM, Maffei VJ, Zaneveld JR et al (2020) PICRUSt2 for prediction of metagenome functions. Nat Biotechnol 38:685–688
Noecker C, Eng A, Srinivasan S et al (2016) Metabolic model-based integration of microbiome taxonomic and metabolomic profiles elucidates mechanistic links between ecological and metabolic variation. MSystems 1:e00013–e00015
Larsen PE, Collart FR, Field D et al (2011) Predicted Relative Metabolomic Turnover (PRMT): determining metabolic turnover from a coastal marine metagenomic dataset. Microb Inform Exp 1:1–11
Bhattacharyya M, Chakrabarti S (2015) Identification of important interacting proteins (IIPs) in plasmodium falciparum using large-scale interaction network analysis and in-silico knock-out studies. Malar J 14:1–17
Yu H, Kim PM, Sprecher E et al (2007) The importance of bottlenecks in protein networks: correlation with gene essentiality and expression dynamics. PLoS Comput Biol 3:713–720
Hagberg A, Swart PS, Chult D (2008) Exploring network structure, dynamics, and function using networkx. N. p. Web, USA
Bag AK, Mandloi S, Jarmalavicius S et al (2019) Connecting signaling and metabolic pathways in EGF receptor-mediated oncogenesis of glioblastoma. PLoS Comput Biol 15:e1007090
Kumar K, Bose S, Chakrabarti S (2021) Identification of cross-pathway connections via protein-protein interactions linked to altered states of metabolic enzymes in cervical cancer. Front Med 8:1949
Biswas N, Kumar K, Bose S et al (2020) Analysis of Pan-Omics Data in Human Interactome Network (APODHIN). Front Genet 11:589231
Biswas N, Chakrabarti S (2020) Artificial Intelligence (AI)-based systems biology approaches in multi-omics data analysis of cancer. Front Oncol 10:588221
Bose S, Kumar K, Chakrabarti S (2021) System biology and network analysis approaches on oxidative stress in cancer. In: Chakraborti S, Ray BK, Roychowdhury S (eds) Handbook of oxidative stress in cancer: mechanistic aspects. Springer, Singapore
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Kumar, K. et al. (2023). Integrating Multi-Omics Data to Construct Reliable Interconnected Models of Signaling, Gene Regulatory, and Metabolic Pathways. In: Nguyen, L.K. (eds) Computational Modeling of Signaling Networks. Methods in Molecular Biology, vol 2634. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-3008-2_6
Download citation
DOI: https://doi.org/10.1007/978-1-0716-3008-2_6
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-3007-5
Online ISBN: 978-1-0716-3008-2
eBook Packages: Springer Protocols