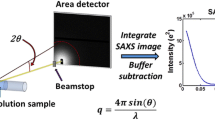
Abstract
The biologically critical, exquisite specificity and efficiency of nucleases, such as those acting in DNA repair and replication, often emerge in the context of multiple other macromolecules. The evolved complexity also makes biologically relevant nuclease assays challenging and low-throughput. Meiotic recombination 11 homolog 1 (MRE11) is an exemplary nuclease that initiates DNA double-strand break (DSB) repair and processes stalled DNA replication forks. Thus, DNA resection by MRE11 nuclease activity is critical for multiple DSB repair pathways as well as in replication. Traditionally, in vitro nuclease activity of purified enzymes is studied either through gel-based assays or fluorescence-based assays like fluorescence resonance energy transfer (FRET). However, adapting these methods for a high-throughput application such as inhibitor screening can be challenging. Gel-based approaches are slow, and FRET assays can suffer from interference and distance limitations. Here we describe an alternative methodology to monitor nuclease activity by measuring the small-angle X-ray scattering (SAXS) interference pattern from gold nanoparticles (Au NPs) conjugated to 5′-ends of dsDNA using X-ray scattering interferometry (XSI). In addition to reporting on the enzyme activity, XSI can provide insight into DNA-protein interactions, aiding in the development of inhibitors that trap enzymes on the DNA substrate. Enabled by efficient access to synchrotron beamlines, sample preparation, and the feasibility of high-throughput XSI data collection and processing pipelines, this method allows for far greater speeds with less sample consumption than conventional SAXS techniques. The reported metrics and methods can be generalized to monitor not only other nucleases but also most other DNA-protein interactions.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Syed A, Tainer JA (2018) The MRE11–RAD50–NBS1 complex conducts the orchestration of damage signaling and outcomes to stress in DNA replication and repair. Annu Rev Biochem 87:263–294. https://doi.org/10.1146/annurev-biochem-062917-012415
Paull TT (2018) 20 years of Mre11 biology: no end in sight. Mol Cell 71:419–427. https://doi.org/10.1016/j.molcel.2018.06.033
Stracker TH, Petrini JHJ (2011) The MRE11 complex: starting from the ends. Nat Rev Mol Cell Biol 12:90–103. https://doi.org/10.1038/nrm3047
Hopfner K-P, Karcher A, Craig L et al (2001) Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell 105:473–485. https://doi.org/10.1016/S0092-8674(01)00335-X
Williams RS, Moncalian G, Williams JS et al (2008) Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell 135:97–109. https://doi.org/10.1016/j.cell.2008.08.017
Park YB, Chae J, Kim YC, Cho Y (2011) Crystal structure of human Mre11: understanding tumorigenic mutations. Structure 19:1591–1602. https://doi.org/10.1016/j.str.2011.09.010
Shibata A, Moiani D, Arvai AS et al (2014) DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol Cell 53:7–18. https://doi.org/10.1016/j.molcel.2013.11.003
Chen C, Hildebrandt N (2020) Resonance energy transfer to gold nanoparticles: NSET defeats FRET. TrAC Trends Anal Chem 123:115748. https://doi.org/10.1016/j.trac.2019.115748
Hura GL, Menon AL, Hammel M et al (2009) Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS). Nat Methods 6:606–612. https://doi.org/10.1038/nmeth.1353
Brosey CA, Tainer JA (2019) Evolving SAXS versatility: solution X-ray scattering for macromolecular architecture, functional landscapes, and integrative structural biology. Curr Opin Struct Biol 58:197–213. https://doi.org/10.1016/j.sbi.2019.04.004
Tang HYH, Tainer JA, Hura GL (2017) High resolution distance distributions determined by X-ray and neutron scattering. In: Chaudhuri B, Muñoz IG, Qian S, Urban VS (eds) Biological small angle scattering: techniques, strategies and tips. Springer, Singapore, pp 167–181
Rambo RP, Tainer JA (2013) Super-resolution in solution X-ray scattering and its applications to structural systems biology. Annu Rev Biophys 42:415–441. https://doi.org/10.1146/annurev-biophys-083012-130301
Rambo RP, Tainer JA (2013) Accurate assessment of mass, models and resolution by small-angle scattering. Nature 496:477–481. https://doi.org/10.1038/nature12070
Putnam CD, Hammel M, Hura GL, Tainer JA (2007) X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys 40:191–285. https://doi.org/10.1017/S0033583507004635
Rambo RP, Tainer JA (2011) Characterizing flexible and instrinsically unstructured biological macromolecules by SAS using the Porod-Debye Law. Biopolymers 95:559–571. https://doi.org/10.1002/bip.21638
Young T (1804) I. The Bakerian Lecture. Experiments and calculations relative to physical optics. Philos Trans R Soc Lond 94:1–16. https://doi.org/10.1098/rstl.1804.0001
Hura GL, Tsai C-L, Claridge SA et al (2013) DNA conformations in mismatch repair probed in solution by X-ray scattering from gold nanocrystals. Proc Natl Acad Sci 110:17308–17313. https://doi.org/10.1073/pnas.1308595110
Vainshtein BK, Feigin LA, Lvov YM et al (1980) Determination of the distance between heavy-atom markers in haemoglobin and histidine decarboxylase in solution by small-angle X-ray scattering. FEBS Lett 116:107–110. https://doi.org/10.1016/0014-5793(80)80539-4
Mathew-Fenn RS, Das R, Silverman JA et al (2008) A molecular ruler for measuring quantitative distance distributions. PLoS One 3:e3229. https://doi.org/10.1371/journal.pone.0003229
Mathew-Fenn RS, Das R, Harbury PAB (2008) Remeasuring the double helix. Science 322:446–449. https://doi.org/10.1126/science.1158881
Mastroianni AJ, Sivak DA, Geissler PL, Alivisatos AP (2009) Probing the conformational distributions of subpersistence length DNA. Biophys J 97:1408–1417. https://doi.org/10.1016/j.bpj.2009.06.031
Shi X, Herschlag D, Harbury PAB (2013) Structural ensemble and microscopic elasticity of freely diffusing DNA by direct measurement of fluctuations. Proc Natl Acad Sci 110:E1444–E1451. https://doi.org/10.1073/pnas.1218830110
Shi X, Beauchamp KA, Harbury PB, Herschlag D (2014) From a structural average to the conformational ensemble of a DNA bulge. Proc Natl Acad Sci 111:E1473–E1480. https://doi.org/10.1073/pnas.1317032111
Shi X, Huang L, Lilley DMJ et al (2016) The solution structural ensembles of RNA kink-turn motifs and their protein complexes. Nat Chem Biol 12:146–152. https://doi.org/10.1038/nchembio.1997
Zettl T, Mathew RS, Shi X et al (2018) Gold nanocrystal labels provide a sequence–to–3D structure map in SAXS reconstructions. Sci Adv 4:eaar4418. https://doi.org/10.1126/sciadv.aar4418
Zettl T, Mathew RS, Seifert S et al (2016) Absolute intramolecular distance measurements with angstrom-resolution using anomalous small-angle X-ray scattering. Nano Lett 16:5353–5357. https://doi.org/10.1021/acs.nanolett.6b01160
Claridge SA, Mastroianni AJ, Au YB et al (2008) Enzymatic ligation creates discrete multinanoparticle building blocks for self-assembly. J Am Chem Soc 130:9598–9605. https://doi.org/10.1021/ja8026746
Tubbs JL, Latypov V, Kanugula S et al (2009) Flipping of alkylated DNA damage bridges base and nucleotide excision repair. Nature 459:808–813. https://doi.org/10.1038/nature08076
Eckelmann BJ, Bacolla A, Wang H et al (2020) XRCC1 promotes replication restart, nascent fork degradation and mutagenic DNA repair in BRCA2-deficient cells. NAR Cancer 2:zcaa013. https://doi.org/10.1093/narcan/zcaa013
Thapar R, Wang JL, Hammel M et al (2020) Mechanism of efficient double-strand break repair by a long non-coding RNA. Nucleic Acids Res 48:10953–10972. https://doi.org/10.1093/nar/gkaa784
Zandarashvili L, Langelier M-F, Velagapudi UK et al (2020) Structural basis for allosteric PARP-1 retention on DNA breaks. Science 368:eaax6367. https://doi.org/10.1126/science.aax6367
Houl JH, Ye Z, Brosey CA et al (2019) Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death. Nat Commun 10:5654. https://doi.org/10.1038/s41467-019-13508-4
Bacolla A, Tainer JA, Vasquez KM, Cooper DN (2016) Translocation and deletion breakpoints in cancer genomes are associated with potential non-B DNA-forming sequences. Nucleic Acids Res 44:5673–5688. https://doi.org/10.1093/nar/gkw261
Bacolla A, Ye Z, Ahmed Z, Tainer JA (2019) Cancer mutational burden is shaped by G4 DNA, replication stress and mitochondrial dysfunction. Prog Biophys Mol Biol 147:47–61. https://doi.org/10.1016/j.pbiomolbio.2019.03.004
Seo SH, Bacolla A, Yoo D et al (2020) Replication-based rearrangements are a common mechanism for SNCA duplication in Parkinson’s disease. Mov Disord 35:868–876. https://doi.org/10.1002/mds.27998
Classen S, Hura GL, Holton JM et al (2013) Implementation and performance of SIBYLS: a dual endstation small-angle X-ray scattering and macromolecular crystallography beamline at the advanced light source. J Appl Crystallogr 46:1–13. https://doi.org/10.1107/S0021889812048698
Dyer KN, Hammel M, Rambo RP et al (2014) High-throughput SAXS for the characterization of biomolecules in solution: a practical approach. Methods Mol Biol 1091:245–258
Glatter O (1977) A new method for the evaluation of small-angle scattering data. J Appl Crystallogr 10:415–421. https://doi.org/10.1107/S0021889877013879
Tully MD, Tarbouriech N, Rambo RP, Hutin S (2021) Analysis of SEC-SAXS data via EFA deconvolution and scatter. J Vis Exp (167):e61578. https://doi.org/10.3791/61578
Hopkins JB, Gillilan RE, Skou S (2017) BioXTAS RAW: improvements to a free open-source program for small-angle X-ray scattering data reduction and analysis. J Appl Crystallogr 50:1545–1553. https://doi.org/10.1107/S1600576717011438
Manalastas-Cantos K, Konarev PV, Hajizadeh NR, et al (2021) ATSAS 3.0: expanded functionality and new tools for small-angle scattering data analysis. J Appl Crystallogr 54:343–355. https://doi.org/10.1107/S1600576720013412
Acknowledgments
The work is supported by R35 CA220430, by the Cancer Prevention Research Institute of Texas (CPRIT-grant#RP180813), and by a Robert A. Welch Chemistry Chair. Efforts to apply SAXS to characterize eukaryotic pathways relevant to human cancers and merge nano- to mesoscale structures are supported in part by National Cancer Institute grants Structural Biology of DNA Repair (SBDR) CA092584. SAXS data was collected at the Advanced Light Source (ALS) beamline SIBYLS which is supported by the DOE-BER IDAT DE-AC02-05CH11231 and NIGMS ALS-ENABLE (P30 GM124169 and S10OD018483).
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Rosenberg, D.J., Syed, A., Tainer, J.A., Hura, G.L. (2022). Monitoring Nuclease Activity by X-Ray Scattering Interferometry Using Gold Nanoparticle-Conjugated DNA. In: Mosammaparast, N. (eds) DNA Damage Responses. Methods in Molecular Biology, vol 2444. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2063-2_12
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2063-2_12
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2062-5
Online ISBN: 978-1-0716-2063-2
eBook Packages: Springer Protocols