Abstract
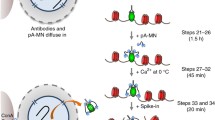
Endonucleolytic cleavage of DNA ends by the human Mre11-Rad50-Nbs1 (MRN) complex occurs in a manner that is promoted by DNA-dependent protein kinase (DNA-PK). A method is described to isolate DNA-PK-bound fragments released from chromatin in human cells using a modified Gentle Lysis and Size Selection chromatin immunoprecipitation (GLASS-ChIP) protocol. This method, combined with real-time PCR or next-generation sequencing, can identify sites of MRN endonucleolytic cutting adjacent to DNA-PK binding sites in human cells.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Pannunzio NR, Watanabe G, Lieber MR (2018) Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J Biol Chem 293:10512–10523. https://doi.org/10.1074/jbc.TM117.000374
Symington LS (2016) Mechanism and regulation of DNA end resection in eukaryotes. Crit Rev Biochem Mol Biol 51:195–212. https://doi.org/10.3109/10409238.2016.1172552
Jasin M, Rothstein R (2013) Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol 5:a012740. https://doi.org/10.1101/cshperspect.a012740
Paull TT (2018) 20 years of Mre11 biology: no end in sight. Mol Cell 71:419–427. https://doi.org/10.1016/j.molcel.2018.06.033
Wright WD, Shah SS, Heyer W-D (2018) Homologous recombination and the repair of DNA double-strand breaks. J Biol Chem 293:10524–10535. https://doi.org/10.1074/jbc.TM118.000372
Blackford AN, Jackson SP (2017) ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol Cell 66:801–817. https://doi.org/10.1016/j.molcel.2017.05.015
Deshpande RA, Myler LR, Soniat MM et al (2020) DNA-dependent protein kinase promotes DNA end processing by MRN and CtIP. Sci Adv 6:eaay0922. https://doi.org/10.1126/sciadv.aay0922
Jones CE, Forsburg SL (2021) Monitoring Schizosaccharomyces pombe genome stress by visualizing end-binding protein Ku. Biology Open 10:bio054346. https://doi.org/10.1242/bio.054346
Kim J-S, Krasieva TB, Kurumizaka H et al (2005) Independent and sequential recruitment of NHEJ and HR factors to DNA damage sites in mammalian cells. J Cell Biol 170:341–347. https://doi.org/10.1083/jcb.200411083
Kochan JA, Desclos ECB, Bosch R et al (2017) Meta-analysis of DNA double-strand break response kinetics. Nucleic Acids Res 45:12625–12637. https://doi.org/10.1093/nar/gkx1128
Wu D, Topper LM, Wilson TE (2008) Recruitment and dissociation of nonhomologous end joining proteins at a DNA double-strand break in Saccharomyces cerevisiae. Genetics 178:1237–1249. https://doi.org/10.1534/genetics.107.083535
Yang G, Liu C, Chen S-H et al (2018) Super-resolution imaging identifies PARP1 and the Ku complex acting as DNA double-strand break sensors. Nucleic Acids Res 46:3446–3457. https://doi.org/10.1093/nar/gky088
Lobrich M, Shibata A, Beucher A et al (2010) gammaH2AX foci analysis for monitoring DNA double-strand break repair: strengths, limitations and optimization. Cell Cycle 9:662–669
Mao Z, Bozzella M, Seluanov A et al (2008) Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair (Amst) 7:1765–1771. https://doi.org/10.1016/j.dnarep.2008.06.018
Iacovoni JS, Caron P, Lassadi I et al (2010) High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J 29:1446–1457. https://doi.org/10.1038/emboj.2010.38
Vítor AC, Huertas P, Legube G et al (2020) Studying DNA double-Strand break repair: an ever-growing toolbox. Front Mol Biosci 7:24. https://doi.org/10.3389/fmolb.2020.00024
Aymard F, Bugler B, Schmidt CK, Guillou E, Caron P, Briois S, Iacovoni JS, Daburon V, Miller KM, Jackson SP, Legube G. (2014) Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat Struct Mol Biol. 21(4):366–374. PMCID: PMC4300393
Zhou Y, Paull TT (2021) Quantifying DNA end resection in human cells. Methods Mol Biol 2153:59–69. https://doi.org/10.1007/978-1-0716-0644-5_5
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Deshpande, R.A., Paull, T.T. (2022). Characterization of DNA-PK-Bound End Fragments Using GLASS-ChIP. In: Mosammaparast, N. (eds) DNA Damage Responses. Methods in Molecular Biology, vol 2444. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2063-2_11
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2063-2_11
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2062-5
Online ISBN: 978-1-0716-2063-2
eBook Packages: Springer Protocols