You have full access to this open access chapter, Download protocol PDF
Similar content being viewed by others
Correction to: Chapter 4 in: Aik T. Ooi (ed.), Single-Cell Protein Analysis: Methods and Protocols, Methods in Molecular Biology, vol. 2386, https://doi.org/10.1007/978-1-0716-1771-7_4
The chapter was inadvertently published with incorrect figure legends, reference citations, and order of references.
These errors have been corrected by updating the correct figure legends, reference citations, and the order of references as seen below:
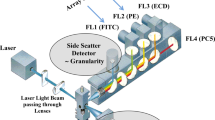
Figure 2 Legend was: Example of population selection for unmixing on single stain and unstained reference controls. Spectral plot (right) shows specific signature of each fluorochrome and cellular autofluorescence
Figure 2 Legend has been corrected as: Anti-IgG antibody cross-reacts with anti-TCR Vd2 (A) and anti-IgA (B). Left plots, protocol with simultaneous staining; right plots, modified protocol with sequential staining
Figure 3 Legend was: Manual Gating Strategy, part 1
Figure 3 Legend has been corrected as: CD19 SparNIR685 signal pre- and post-fixation step. Post-fixation staining shows better resolution of CD19+ cells
Figure 4 Legend was: Manual Gating Strategy, part 2
Figure 4 Legend has been corrected as: Example of population selection for unmixing on single stain and unstained reference controls. Spectral plot (right) shows specific signature of each fluorochrome and cellular autofluorescence
Figure 5 Legend was: Manual Gating Strategy, part 3
Figure 5 Legend has been corrected as: Manual Gating Strategy, part 1
Figure 6 Legend was: Visualization of high-dimensional data using t-SNE and UMAP algorithms on PBMC -excluding monocytes- from three concatenated samples. A. Biaxial plot with t-SNE (upper) and UMAP (lower) maps. B. Overlayed color-coded expression of several markers (CD3, CD4, CD8, TIGIT, CD57, CD19, IgM, IgD, CXCR5, CCR6, CD56, CD16 CD161, CD123 and CD27) on dimensional reduction t-SNE map. Data generated with the OMIQ Platform (https://omiq.ai)
Figure 6 Legend has been corrected as: Manual Gating Strategy, part 2
Figure 7 Legend was: Anti-IgG antibody cross-reacts with anti-TCR Vd2 (A) and anti-IgA (B). Left plots, standard protocol with simultaneous staining; right plots, alternative staining with sequential staining
Figure 7 Legend has been corrected as: Manual Gating Strategy, part 3
Figure 8 Legend was: CD19 SparNIR685 signal pre- and post-fixation step. Post-fixation staining shows better resolution of CD19+ cells
Figure 8 Legend has been corrected as: Visualization of high-dimensional data using t-SNE and UMAP algorithms on PBMC—excluding monocytes—from three concatenated samples. (A) Biaxial plot with t-SNE (upper) and UMAP (lower) maps. (B) Overlayed color-coded expression of several markers (CD3, CD4, CD8, TIGIT, CD57, CD19, IgM, IgD, CXCR5, CCR6, CD56, CD16, CD161, CD123, and CD27) on dimensional reduction t-SNE map. Data generated with the OMIQ Platform (https://omiq.ai)
The following citations have been revised:
Section 3 : Methods
[8] has been updated as (see Notes 9 and 10)
[9] has been updated as [8]
[10] has been updated as [9]
[8, 11] has been updated as [10, 11]
Section 4 Notes
3. [14] has been updated as [13]
5. [15] has been updated as [14]
7. [9,16] has been update as [8, 15]
9. [17] has been updated as [16]
10. [17] has been updated as [16]
11. [15, 18,19] has been updated as [17–19]
References have been sorted in the following order in the updated version of the chapter.
-
1.
Herzenberg LA, Sweet RG, Herzenberg LA (1976) Fluorescence-activated cell sorting. Sci Am 234(3):108–117. https://doi.org/10.1038/scientificamerican0376-108
-
2.
Biolegend History of Flow Cytometry. https://www.biolegend.com/en-us/history-of-flow
-
3.
Herzenberg LA, Parks D, Sahaf B, et al (2002) The history and future of the fluorescence activated cell sorter and flow cytometry: a view from Stanford. Clin Chem 48:1819–1827. https://doi.org/10.1093/clinchem/48.10.1819
-
4.
Hu B, Guo H, Zhou P, Shi ZL (2020) Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol 19(3):141–154. https://doi.org/10.1038/s41579-020-00459-7
-
5.
Nicola M, Alsafi Z, Sohrabi C, et al (2020) The socio-economic implications of the coronavirus pandemic (COVID-19): a review. Int J Surg 78:185–193
-
6.
John Hopkins Coronavirus Resource Center
-
7.
Nolan JP, Condello D (2013) Spectral flow cytometry. Curr Protoc Cytom. https://doi.org/10.1002/0471142956.cy0127s63
-
8.
Mahnke YD, Roederer M (2007) Optimizing a multicolor immunophenotyping assay. Clin Lab Med 27(3):469-485, v
-
9.
Hulspas R (2010) Titration of fluorochrome-conjugated antibodies for labeling cell surface markers on live cells. Curr Protoc Cytom. https://doi.org/10.1002/0471142956.cy0629s54
-
10.
Niewold P, Ashhurst TM, Smith AL, King NJC (2020) Evaluating spectral cytometry for immune profiling in viral disease. Cytom Part A 97(11):1165–1179. https://doi.org/10.1002/cyto.a.24211
-
11.
Schmutz S, Valente M, Cumano A, Novault S (2016) Spectral cytometry has unique properties allowing multicolor analysis of cell suspensions isolated from solid tissues. PLoS One 11(8):e0159961. https://doi.org/10.1371/journal.pone.0159961
-
12.
Interim Laboratory Biosafety Guidelines for Handling and Processing Specimens Associated with Coronavirus Disease 2019 (COVID-19). Centers Dis Control Prevention
-
13.
Zhao Q, Meng M, Kumar R, et al (2020) Lymphopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: a systemic review and meta-analysis. Int J Infect Dis 96:131–135. https://doi.org/10.1016/j.ijid.2020.04.086
-
14.
Jalbert E, Shikuma CM, Ndhlovu LC, Barbour JD (2013) Sequential staining improves detection of CCR2 and CX3CR1 on monocytes when simultaneously evaluating CCR5 by multicolor flow cytometry. Cytometry A 83(3):280–286.. https://doi.org/10.1002/cyto.a.22257
-
15.
Ferrer-Font L, Pellefigues C, Mayer JU, et al (2020) Panel design and optimization for high-dimensional immunophenotyping assays using spectral flow cytometry. Curr Protoc Cytom 92:1–25. https://doi.org/10.1002/cpcy.70
-
16.
Park LM, Lannigan J, Jaimes MC (2020) Forty-color full spectrum flow cytometry panel for deep immunophenotyping of major cell subsets in human peripheral blood. Cytometry A 97:1044–1051. https://doi.org/10.1002/cyto.a.24213
-
17.
Palit S, Heuser C, De Almeida GP, et al (2019) Meeting the challenges of high-dimensional single-cell data analysis in immunology. Front Immunol 10:1515. https://doi.org/10.3389/fimmu.2019.01515
-
18.
Becht E, McInnes L, Healy J, et al (2019) Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. https://doi.org/10.1038/nbt.4314
-
19.
Amir E ad D, Lee B, Badoual P, et al (2019) Development of a comprehensive antibody staining database using a standardized analytics pipeline. Front Immunol 10:1315. https://doi.org/10.3389/fimmu.2019.01315
-
20.
Melsen JE, van Ostaijen-ten Dam MM, Lankester AC, et al (2020) A comprehensive workflow for applying single-cell clustering and pseudotime analysis to flow cytometry data. J Immunol 205(3):864–871. https://doi.org/10.4049/jimmunol.1901530
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Fernandez, M.A., Alzayat, H., Jaimes, M.C., Kharraz, Y., Requena, G., Mendez, P. (2022). Correction to: High-Dimensional Immunophenotyping with 37-Color Panel Using Full-Spectrum Cytometry. In: Ooi, A.T. (eds) Single-Cell Protein Analysis. Methods in Molecular Biology, vol 2386. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-1771-7_18
Download citation
DOI: https://doi.org/10.1007/978-1-0716-1771-7_18
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-1770-0
Online ISBN: 978-1-0716-1771-7
eBook Packages: Springer Protocols