Abstract
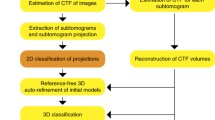
Cryo-electron microscopy has established as a mature structural biology technique to elucidate the three-dimensional structure of biological macromolecules. The Coulomb potential of the sample is imaged by an electron beam, and fast semi-conductor detectors produce movies of the sample under study. These movies have to be further processed by a whole pipeline of image-processing algorithms that produce the final structure of the macromolecule. In this chapter, we illustrate this whole processing pipeline putting in value the strength of “meta algorithms,” which are the combination of several algorithms, each one with different mathematical rationale, in order to distinguish correctly from incorrectly estimated parameters. We show how this strategy leads to superior performance of the whole pipeline as well as more confident assessments about the reconstructed structures. The “meta algorithms” strategy is common to many fields and, in particular, it has provided excellent results in bioinformatics. We illustrate this combination using the workflow engine, Scipion.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Benjin X, Ling L (2020) Developments, applications, and prospects of cryo-electron microscopy. Protein Sci 29:872–882
Lyumkis D (2019) Challenges and opportunities in cryo-em single-particle analysis. J Biol Chem 294:5181–5197
Eisenstein M (2018) Drug designers embrace cryo-EM. Nat Biotechnol 36:557–558
Scapin G, Potter CS, Carragher B (2018) Cryo-em for small molecules discovery, design, understanding, and application. Cell Chem Biol 25:1318–1325
Saur M, Hartshorn MJ, Dong J, Reeks J et al (2019) Fragment-based drug discovery using cryo-em Drug discovery today. doi: https://doi.org/10.1016/j.drudis.2019.12.006
Jonic S (2017) Computational methods for analyzing conformational variability of macromolecular complexes from cryo-electron microscopy images. Curr Opin Struct Biol 43:114–121
Sorzano COS, Jiménez A, Mota J, Vilas JL et al (2019) Survey of the analysis of continuous conformational variability of biological macromolecules by electron microscopy. Acta Crystallogr Sect F, Struct Biol Commun 75:19–32
Arnold SA, Müller SA, Schmidli C et al (2018) Miniaturizing EM sample preparation: opportunities, challenges, and “visual proteomics”. Proteomics 18:e1700176
Faruqi AR, McMullan G (2018) Direct imaging detectors for electron microscopy. Nucl Instrum Methods Phys Res, Sect A 878:180–190
Vilas JL, Gómez-Blanco J, Conesa P et al (2018) MonoRes: automatic and unbiased estimation of local resolution for electron microscopy maps. Structure 26:337–344
de la Rosa-Trevín JM, Quintana A, Del Cano L et al (2016) Scipion: a software framework toward integration, reproducibility and validation in 3D electron microscopy. J Struct Biol 195:93–99
Wang Z, Hryc CF, Bammes B et al (2014) An atomic model of brome mosaic virus using direct electron detection and real-space optimization. Nat Commun 5:4808
Heymann JB, Marabini R, Kazemi M et al (2018) The first single particle analysis map challenge: a summary of the assessments. J Struct Biol 204:291–300
Sorzano COS, Vargas J, Oton J et al (2017) A review of resolution measures and related aspects in 3D electron microscopy. Prog Biophys Mol Biol 124:1–30
Vilas JL, Tagare HD, Vargas J et al (2020) Measuring local-directional resolution and local anisotropy in cryo-EM maps. Nat Commun 11:55
Ramírez-Aportela E, Mota J, Conesa P et al (2019) Deep-res: a new deep-learning- and aspect-based local resolution method for electron-microscopy maps. IUCRj 6:1054–1063
Sorzano COS, Fernández-Giménez E, Peredo-Robinson V et al (2018) Blind estimation of DED camera gain in electron microscopy. J Struct Biol 203:90–93
Li X, Mooney P, Zheng S, Booth CR et al (2013) Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods 10:584–590
Zheng SQ, Palovcak E, Armache JP et al (2017) Motion- cor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14:331–332
Abrishami V, Vargas J, Li X, Cheng Y et al (2015) Alignment of direct detection device micrographs using a robust optical flow approach. J Struct Biol 189:163–176
Tegunov D, Cramer P (2019) Real-time cryo-electron microscopy data preprocessing with warp. Nat Methods 16:1146–1152
de la Rosa-Trevín JM, Otón J, Marabini R et al (2013) Xmipp 3.0: an improved software suite for image processing in electron microscopy. J Struct Biol 184(2):321–328
Sorzano COS, Jonic S, Núñez Ramírez R et al (2007) Fast, robust and accurate determination of transmission electron microscopy contrast transfer function. J Struct Biol 160:249–262
Zhang K (2016) Gctf: real-time ctf determination and correction. J Struct Biol 193:1–12
Rohou A, Grigorieff N (2015) Ctffind4: fast and accurate defocus estimation from electron micrographs. J Struct Biol 192:216–221
Maluenda D, Majtner T, Horvath P et al (2019) Flexible workflows for on-the-fly electron-microscopy single-particle image processing using scipion. Acta Crystallogr Sect D, Struct Biol 75:882–894
Marabini R, Carragher B, Chen S et al (2015) Ctf challenge: result summary. J Struct Biol 190:348–359
Voss NR, Yoshioka CK, Radermacher M et al (2009) Dog picker and tiltpicker: software tools to facilitate particle selection in single particle electron microscopy. J Struct Biol 166(2):205–213
Scheres SHW (2015) Semi-automated selection of cryo-em particles in relion-1.3. J Struct Biol 189:114–122
Abrishami V, Zaldívar-Peraza A, de la Rosa-Trevín JM et al (2013) A pattern matching approach to the automatic selection of particles from low-contrast electron micrographs. Bioinformatics 29:2460–2468
Bepler T, Morin A, Rapp M, Brasch J et al (2019) Topaz: a positive-unlabeled convolutional neural network cryoem particle picker that can pick any size and shape particle. Microsc Microanal 25:986–987
Wagner T, Merino F, Stabrin M et al (2019) Sphire-cryolo is a fast and accurate fully automated particle picker for cryo-EM. Commun Biol 2:218
Sanchez-Garcia R, Segura J, Maluenda D et al (2018) Deep consensus, a deep learning-based approach for particle pruning in cryo-electron microscopy. IUCrJ 5:854–865
Sánchez-García R, Segura J, Maluenda D et al (2020) Micrograph cleaner: a python package for cryo-EM micrograph cleaning using deep learning. bioRxiv. https://doi.org/10.1101/677542
Vargas J, Abrishami V, Marabini R et al (2013) Particle quality assessment and sorting for automatic and semiautomatic particle-picking techniques. J Struct Biol 183:342–353
Punjani A, Brubaker MA, Fleet DJ (2017) Building proteins in a day: efficient 3D molecular structure estimation with electron cryomicroscopy. IEEE Trans Pattern Anal Mach Intell 39:706–718
Sorzano COS, Bilbao-Castro JR, Shkolnisky Y et al (2010) A clustering approach to multireference alignment of single-particle projections in electron microscopy. J Struct Biol 171:197–206
Sorzano COS, Vargas J, de la Rosa-Trevín JM et al (2014) Outlier detection for single particle analysis in electron microscopy. In: Proc. Intl. Work-Conference on Bioinformatics and Biomedical Engineering, IWBBIO, p 950
Vargas J, Álvarez-Cabrera AL, Marabini R et al (2014) Efficient initial volume determination from electron microscopy images of single particles. Bioinformatics 30:2891–2898
Sorzano COS, Vargas J, de la Rosa-Trevín JM et al (2015) A statistical approach to the initial volume problem in single particle analysis by electron microscopy. J Struct Biol 189:213–219
Scheres SHW (2012) Relion: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180:519–530
Tang G, Peng L, Baldwin PR, Mann DS et al (2007) Eman2: an extensible image processing suite for electron microscopy. J Struct Biol 157:38–46
Reboul CF, Eager M, Elmlund D, Elmlund H (2018) Single-particle cryo-EM- improved ab initio 3D reconstruction with simple/prime. Protein Sci 27:51–61
Sorzano COS, Vargas J, Vilas JL et al (2018) Swarm optimization as a consensus technique for electron microscopy initial volume. Appl Anal Optim 2:299–313
Gomez-Blanco J, Kaur S, Ortega J, Vargas J (2019) A robust approach to ab initio cryo-electron microscopy initial volume determination. J Struct Biol 208:107397
Jimenez A, Jonic S, Majtner T et al (2019) Validation of electron microscopy initial models via small angle x-ray scattering curves. Bioinformatics 35:2427–2433
Kimanius D, Forsberg BO, Scheres SH, Lindahl E (2016) Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. elife 5:e18722
Sorzano COS, Marabini R, Vargas J et al (2014) Computational Methods for Three-Dimensional Microscopy Reconstruction, Springer, chap Interchanging geometry information in electron microscopy single particle analysis: mathematical context for the development of a standard, pp 7–42
Cardone G, Heymann JB, Steven AC (2013) One number does not fit all: mapping local variations in resolution in cryo-em reconstructions. J Struct Biol 184:226–236
Kucukelbir A, Sigworth FJ, Tagare HD (2014) Quantifying the local resolution of cryo-EM density maps. Nat Methods 11:63–65
Ramírez-Aportela E, Vilas JL, Glukhova A et al (2019) Automatic local resolution-based sharpening of cryo-EM maps. Bioinformatics 36:765–772
Fernández JJ, Luque D, Castón JR, Carrascosa JL (2008) Sharpening high resolution information in single particle electron cryomicroscopy. J Struct Biol 164(1):170–175
Vilas JL, Vargas J, Martínez M et al (2020b) Re-examining the spectra of macromolecules: current practice of spectral quasi b-factor flattening. J Struct Biol 209:107447
Jakobi AJ, Wilmanns M, Sachse C (2017) Model-based local density sharpening of cryo-EM maps. elife 6:e27131
Naydenova K, Russo CJ (2017) Measuring the effects of particle orientation to improve the efficiency of electron cryomicroscopy. Nat Commun 8:629
Vargas J, Otón J, Marabini R et al (2016) Particle alignment reliability in single particle electron cryomicroscopy: a general approach. Sci Rep 6:21626
Vargas J, Melero R, Gómez-Blanco J et al (2017) Quantitative analysis of 3D alignment quality: its impact on soft-validation, particle pruning and homogeneity analysis. Sci Rep 7:6307
Stagg SM, Noble AJ, Spilman M, Chapman MS (2014) Reslog plots as an empirical metric of the quality of cryo-em reconstructions. J Struct Biol 185:418–426
Heymann B (2015) Validation of 3dem reconstructions: the phantom in the noise. AIMS Biophys 2:21–35
Beckers M, Jakobi AJ, Sachse C (2019) Thresholding of cryo-em density maps by false discovery rate control. IUCrJ 6(1):18–33
Martínez M, Jiménez-Moreno A, Maluenda D et al (2020) Integration of cryo-EM model building software in Scipion. J Chem Inf Model 26:2533–2540
Afonine PV, Klaholz BP, Moriarty NW et al (2018) New tools for the analysis and validation of cryo-em maps and atomic models. Acta Crystallogr Sect D, Struct Biol 74:814–840
Patwardhan A (2017) Trends in the electron microscopy data bank (emdb). Acta Crystallogr Sect D: Struct Biol 73:503–508
Iudin A, Korir PK, Salavert-Torres J et al (2016) Empiar: a public archive for raw electron microscopy image data. Nat Methods 13:387–388
Acknowledgments
The authors would like to acknowledge economical support from: The Spanish Ministry of Economy and Competitiveness through Grants BIO2016-76400-R(AEI/FEDER, UE), the “Comunidad Autónoma de Madrid” through Grant: S2017/BMD-3817. Instituto de Salud Carlos III through Grant: PT17/0009/0010 (ISCIII-SGEFI / ERDF). European Union (EU) and Horizon 2020 through grants: CORBEL (INFRADEV-1-2014-1, Proposal: 654248) Instruct ULTRA (Proposal: 731005), EOSC Life (Proposal: 824087), HighResCells (Proposal: 810057), IMpaCT (Proposal: 857203), EOSC—Synergy (Proposal: 857647), iNEXT-Discovery (Proposal: 871037), and European Regional Development Fund-Project “CERIT Scientific Cloud” (No. CZ.02.1.01/0.0/0.0/16_013/0001802). The authors acknowledge the support and the use of resources of Instruct, a Landmark ESFRI project.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Sorzano, C.O.S. et al. (2021). Image Processing in Cryo-Electron Microscopy of Single Particles: The Power of Combining Methods. In: Owens, R.J. (eds) Structural Proteomics. Methods in Molecular Biology, vol 2305. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-1406-8_13
Download citation
DOI: https://doi.org/10.1007/978-1-0716-1406-8_13
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-1405-1
Online ISBN: 978-1-0716-1406-8
eBook Packages: Springer Protocols