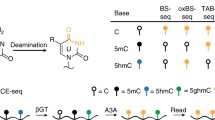
Abstract
5-Methylcytosine (5mC) is one of the most abundant and well-studied chemical DNA modifications of vertebrate genomes. 5mC plays an essential role in genome regulation including: silencing of retroelements, X chromosome inactivation, and heterochromatin stability. Furthermore, 5mC shapes the activity of cis-regulatory elements crucial for cell fate determination. TET enzymes can oxidize 5mC to form 5-hydroxymethylcytosine (5hmC), thereby adding an additional layer of complexity to the DNA methylation landscape dynamics. The advent of techniques enabling genome-wide 5hmC profiling provided critical insights into its genomic distribution, scope, and function. These methods include immunoprecipitation, chemical labeling and capture-based approaches, as well as single-nucleotide 5hmC profiling techniques such as TET-assisted bisulfite sequencing (TAB-seq) and APOBEC-coupled epigenetic sequencing (ACE-seq). Here we provide a detailed protocol for computational analysis required for the genomic alignment of TAB-seq and ACE-seq data, 5hmC calling, and statistical analysis.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Schubeler D (2015) Function and information content of DNA methylation. Nature 517:321–326
Clark SJ, Harrison J, Paul CL et al (1994) High sensitivity mapping of methylated cytosines. Nucleic Acids Res 22:2990–2997
Lister R, Pelizzola M, Dowen RH et al (2009) Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462:315–322
Stadler MB, Murr R, Burger L et al (2011) DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480:490–495
Lister R, Mukamel EA, Nery JR et al (2013) Global epigenomic reconfiguration during mammalian brain development. Science 341:1237905
Bogdanovic O, Smits AH, de la Calle Mustienes E et al (2016) Active DNA demethylation at enhancers during the vertebrate phylotypic period. Nat Genet 48:417–426
Skvortsova K, Tarbashevich K, Stehling M et al (2019) Retention of paternal DNA methylome in the developing zebrafish germline. Nat Commun 10:3054
Tahiliani M, Koh KP, Shen Y et al (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324:930–935
Ito S, Shen L, Dai Q et al (2011) Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333:1300–1303
Mellen M, Ayata P, Heintz N (2017) 5-hydroxymethylcytosine accumulation in postmitotic neurons results in functional demethylation of expressed genes. Proc Natl Acad Sci U S A 114:E7812–E7821
Xu Y, Wu F, Tan L et al (2011) Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell 42:451–464
Wu H, D’Alessio AC, Ito S et al (2011) Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev 25:679–684
Yu M, Hon GC, Szulwach KE et al (2012) Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149:1368–1380
Schutsky EK, DeNizio JE, Hu P et al (2018) Nondestructive, base-resolution sequencing of 5-hydroxymethylcytosine using a DNA deaminase. Nat Biotechnol. https://doi.org/10.1038/nbt.4204
Tanaka K, Okamoto A (2007) Degradation of DNA by bisulfite treatment. Bioorg Med Chem Lett 17:1912–1915
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Li H, Handsaker B, Wysoker A et al (2009) The sequence alignment/Map format and SAMtools. Bioinformatics 25:2078–2079
Chen H, Smith AD, Chen T (2016) WALT: fast and accurate read mapping for bisulfite sequencing. Bioinformatics 32:3507–3509
Robinson JT, Thorvaldsdottir H, Winckler W et al (2011) Integrative genomics viewer. Nat Biotechnol 29:24–26
Andrews S (2010) FastQC: A quality control tool for high throughput sequence data [Online]. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Kriaucionis S, Heintz N (2009) The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324:929–930
Li X, Liu Y, Salz T et al (2016) Whole-genome analysis of the methylome and hydroxymethylome in normal and malignant lung and liver. Genome Res 26:1730–1741
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Skvortsova, K., Bogdanovic, O. (2021). TAB-seq and ACE-seq Data Processing for Genome-Wide DNA hydroxymethylation Profiling. In: Bogdanovic, O., Vermeulen, M. (eds) TET Proteins and DNA Demethylation. Methods in Molecular Biology, vol 2272. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-1294-1_9
Download citation
DOI: https://doi.org/10.1007/978-1-0716-1294-1_9
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-1293-4
Online ISBN: 978-1-0716-1294-1
eBook Packages: Springer Protocols