Abstract
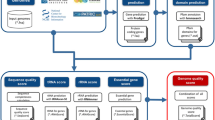
Genome assembly and annotation are two of the key actions that must be undertaken in order to explore the genomic repertoire of (bifido)bacteria. The gathered information can be employed to genomically characterize a given microorganism, and can also be used to perform comparative genome analysis by including other sequenced (bifido)bacterial strains. Here, we highlight various bioinformatic programs able to manage next generation sequencing data starting from the assembly of a genome to the comparative analyses between strains.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM et al (1995) Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496–512
Slatko BE, Gardner AF, Ausubel FM (2018) Overview of next-generation sequencing technologies. Curr Protoc Mol Biol 122:e59
Turroni F, Milani C, Duranti S, Ferrario C, Lugli GA, Mancabelli L, van Sinderen D, Ventura M (2018) Bifidobacteria and the infant gut: an example of co-evolution and natural selection. Cell Mol Life Sci 75:103–118
Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, Belzer C, Delgado Palacio S, Arboleya Montes S, Mancabelli L, Lugli GA, Rodriguez JM, Bode L, de Vos W, Gueimonde M, Margolles A, van Sinderen D, Ventura M (2017) The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev 81:e00036-17
Lugli GA, Milani C, Duranti S, Mancabelli L, Mangifesta M, Turroni F, Viappiani A, van Sinderen D, Ventura M (2018) Tracking the taxonomy of the genus Bifidobacterium based on a Phylogenomic approach. Appl Environ Microbiol 84:e02249-17
Lugli GA, Milani C, Turroni F, Duranti S, Mancabelli L, Mangifesta M, Ferrario C, Modesto M, Mattarelli P, Jiri K, van Sinderen D, Ventura M (2017) Comparative genomic and phylogenomic analyses of the Bifidobacteriaceae family. BMC Genomics 18:568
Lugli GA, Milani C, Turroni F, Duranti S, Ferrario C, Viappiani A, Mancabelli L, Mangifesta M, Taminiau B, Delcenserie V, van Sinderen D, Ventura M (2014) Investigation of the evolutionary development of the genus Bifidobacterium by comparative genomics. Appl Environ Microbiol 80:6383–6394
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477
Darling AC, Mau B, Blattner FR, Perna NT (2004) Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403
Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ (2010) Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964
Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW (2007) RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35:3100–3108
O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, Astashyn A, Badretdin A, Bao Y, Blinkova O, Brover V, Chetvernin V, Choi J, Cox E, Ermolaeva O, Farrell CM, Goldfarb T, Gupta T, Haft D, Hatcher E, Hlavina W, Joardar VS, Kodali VK, Li W, Maglott D, Masterson P, McGarvey KM, Murphy MR, O'Neill K, Pujar S, Rangwala SH, Rausch D, Riddick LD, Schoch C, Shkeda A, Storz SS, Sun H, Thibaud-Nissen F, Tolstoy I, Tully RE, Vatsan AR, Wallin C, Webb D, Wu W, Landrum MJ, Kimchi A et al (2016) Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44:D733–D745
Zhao Y, Tang H, Ye Y (2012) RAPSearch2: a fast and memory-efficient protein similarity search tool for next-generation sequencing data. Bioinformatics 28:125–126
Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A (2016) The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285
Eddy SR (2011) Accelerated profile HMM searches. PLoS Comput Biol 7:e1002195
Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA (2012) Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28:464–469
Zhao Y, Wu J, Yang J, Sun S, Xiao J, Yu J (2012) PGAP: pan-genomes analysis pipeline. Bioinformatics 28:416–418
Pearson WR (2014) BLAST and FASTA similarity searching for multiple sequence alignment. Methods Mol Biol 1079:75–101
Enright AJ, Van Dongen S, Ouzounis CA (2002) An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30:1575–1584
Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, Rattei T, Mende DR, Sunagawa S, Kuhn M, Jensen LJ, von Mering C, Bork P (2016) eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res 44:D286–D293
Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495
Caspi R, Billington R, Ferrer L, Foerster H, Fulcher CA, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Mueller LA, Ong Q, Paley S, Subhraveti P, Weaver DS, Karp PD (2016) The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 44:D471–D480
Richter M, Rossello-Mora R, Oliver Glockner F, Peplies J (2016) JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32:929–931
Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL (2004) Versatile and open software for comparing large genomes. Genome Biol 5:R12
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, Deboy RT, Davidsen TM, Mora M, Scarselli M, Margarit y Ros I, Peterson JD, Hauser CR, Sundaram JP, Nelson WC, Madupu R, Brinkac LM, Dodson RJ, Rosovitz MJ, Sullivan SA, Daugherty SC, Haft DH, Selengut J, Gwinn ML, Zhou L, Zafar N, Khouri H, Radune D, Dimitrov G, Watkins K, O'Connor KJ, Smith S, Utterback TR, White O, Rubens CE, Grandi G, Madoff LC, Kasper DL, Telford JL, Wessels MR, Rappuoli R, Fraser CM (2005) Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial "pan-genome". Proc Natl Acad Sci U S A 102:13950–13955
Richter M, Rossello-Mora R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A 106:19126–19131
Lugli GA, Milani C, Mancabelli L, van Sinderen D, Ventura M (2016) MEGAnnotator: a user-friendly pipeline for microbial genomes assembly and annotation. FEMS Microbiol Lett 363
Duranti S, Mangifesta M, Lugli GA, Turroni F, Anzalone R, Milani C, Mancabelli L, Ossiprandi MC, Ventura M (2017) Bifidobacterium vansinderenii sp. nov., isolated from faeces of emperor tamarin (Saguinus imperator). Int J Syst Evol Microbiol 67(10):3987–3995. https://doi.org/10.1099/ijsem.0.002243
Lugli GA, Mangifesta M, Duranti S, Anzalone R, Milani C, Mancabelli L, Alessandri G, Turroni F, Ossiprandi MC, van Sinderen D, Ventura M (2018) Phylogenetic classification of six novel species belonging to the genus Bifidobacterium comprising Bifidobacterium anseris sp. nov., Bifidobacterium criceti sp. nov., Bifidobacterium imperatoris sp. nov., Bifidobacterium italicum sp. nov., Bifidobacterium margollesii sp. nov. and Bifidobacterium parmae sp. nov. Syst Appl Microbiol 41:173–183
Duranti S, Lugli GA, Napoli S, Anzalone R, Milani C, Mancabelli L, Alessandri G, Turroni F, Ossiprandi MC, van Sinderen D, Ventura M (2019) Characterization of the phylogenetic diversity of five novel species belonging to the genus Bifidobacterium: Bifidobacterium castoris sp. nov., Bifidobacterium callimiconis sp. nov., Bifidobacterium goeldii sp. nov., Bifidobacterium samirii sp. nov. and Bifidobacterium dolichotidis sp. nov. Int J Syst Evol Microbiol 69(5):1288–1298. https://doi.org/10.1099/ijsem.0.003306
Lugli GA, Mancino W, Milani C, Duranti S, Mancabelli L, Napoli S, Mangifesta M, Viappiani A, Anzalone R, Longhi G, van Sinderen D, Ventura M, Turroni F (2019) Dissecting the evolutionary development of the species Bifidobacterium animalis through comparative genomics analyses. Appl Environ Microbiol 85(7):e02806–e02818
Lugli GA, Duranti S, Albert K, Mancabelli L, Napoli S, Viappiani A, Anzalone R, Longhi G, Milani C, Turroni F, Alessandri G, Sela DA, van Sinderen D, Ventura M (2019) Unveiling genomic diversity among members of the species Bifidobacterium pseudolongum, a widely distributed gut commensal of the animal kingdom. Appl Environ Microbiol 85(8):e03065–e03018
Duranti S, Milani C, Lugli GA, Mancabelli L, Turroni F, Ferrario C, Mangifesta M, Viappiani A, Sanchez B, Margolles A, van Sinderen D, Ventura M (2016) Evaluation of genetic diversity among strains of the human gut commensal Bifidobacterium adolescentis. Sci Rep 6:23971
Duranti S, Milani C, Lugli GA, Turroni F, Mancabelli L, Sanchez B, Ferrario C, Viappiani A, Mangifesta M, Mancino W, Gueimonde M, Margolles A, van Sinderen D, Ventura M (2015) Insights from genomes of representatives of the human gut commensal Bifidobacterium bifidum. Environ Microbiol 17:2515–2531
Lugli GA, Milani C, Duranti S, Alessandri G, Turroni F, Mancabelli L, Tatoni D, Ossiprandi MC, van Sinderen D, Ventura M (2019) Isolation of novel gut bifidobacteria using a combination of metagenomic and cultivation approaches. Genome Biol 20:96
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Lugli, G.A. (2021). Assembly, Annotation, and Comparative Analysis of Bifidobacterial Genomes. In: van Sinderen, D., Ventura, M. (eds) Bifidobacteria. Methods in Molecular Biology, vol 2278. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-1274-3_4
Download citation
DOI: https://doi.org/10.1007/978-1-0716-1274-3_4
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-1273-6
Online ISBN: 978-1-0716-1274-3
eBook Packages: Springer Protocols