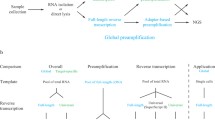
Abstract
A workflow is described for assaying the expression of G protein-coupled receptors (GPCRs) in cultured cells, using a combination of methods that assess GPCR mRNAs. Beginning from the isolation of cDNA and preparation of mRNA, we provide protocols for designing and testing qPCR primers, assaying mRNA expression using qPCR and high-throughput analysis of GPCR mRNA expression via TaqMan qPCR-based, GPCR-selective arrays. We also provide a workflow for analysis of expression from RNA-sequencing (RNA-seq) assays, which can be queried to yield expression of GPCRs and related genes in samples of interest, as well as to test changes in expression between groups, such as in cells treated with drugs or from healthy and diseased subjects. We place priority on optimized protocols that distinguish signal from noise, as GPCR mRNAs are typically present in low abundance, necessitating techniques that maximize sensitivity while minimizing noise. These methods may also be applicable for assessing the expression of members of families of other low abundance genes via high-throughput analyses of mRNAs, followed by independent confirmation and validation of results via qPCR.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Sriram K, Insel PA (2018) G protein-coupled receptors as targets for approved drugs: how many targets and how many drugs? Mol Pharmacol 93:251–258
Alexander SP, Christopoulos A, Davenport AP, Kelly E, Mathie A, Peters JA, Veale EL, Armstrong JF, Faccenda E, Harding SD, Pawson AJ (2019) The concise guide to PHARMACOLOGY 2019/20: G protein-coupled receptors. Br J Pharmacol 176:S21–S141
Sriram K, Moyung K, Corriden R, Carter H, Insel PA (2019) GPCRs show widespread differential mRNA expression and frequent mutation and copy number variation in solid tumor. PLoS Biol 17:e3000434
Sriram K, Wiley SZ, Moyung K, Gorr MW, Salmerón C, Marucut J, French RP, Lowy AM, Insel PA (2019) Detection and quantification of GPCR mRNA: an assessment and implications of data from high-content methods. ACS Omega 4:17048–17059
Insel PA, Sriram K, Gorr MW, Wiley SZ, Michkov A, Salmerón C, Chinn AM (2019) GPCRomics: an approach to discover GPCR drug targets. Trends Pharmacol Sci 40:378–387
Sriram K, Insel PA (2020) A hypothesis for pathobiology and treatment of COVID-19: the centrality of ACE1/ACE2 imbalance. Br J Pharmacol. https://doi.org/10.1111/bph.15082
Bray NL, Pimentel H, Melsted P, Pachter L (2016) Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34:525–527
Soneson C, Love MI, Robinson MD (2015) Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4:1521
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140
Durinck S, Spellman P, Birney E, Huber W (2009) Mapping identifiers for the integration of genomic datasets with the R/bioconductor package biomaRt. Nat Protoc 4:1184–1191
Gupta SK, Haigh BJ, Griffin FJ, Wheeler TT (2013) The mammalian secreted RNases: mechanisms of action in host defence. Innate Immun 19:86–97
Acknowledgements
Support was provided by Academic Senate of the University of California, San Diego and NIH grant RO1A1093957.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Sriram, K., Salmerón, C., Di Nardo, A., Insel, P.A. (2021). Detection of GPCR mRNA Expression in Primary Cells Via qPCR, Microarrays, and RNA-Sequencing. In: Martins, S.A.M., Prazeres, D.M.F. (eds) G Protein-Coupled Receptor Screening Assays. Methods in Molecular Biology, vol 2268. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-1221-7_2
Download citation
DOI: https://doi.org/10.1007/978-1-0716-1221-7_2
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-1220-0
Online ISBN: 978-1-0716-1221-7
eBook Packages: Springer Protocols