Abstract
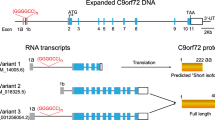
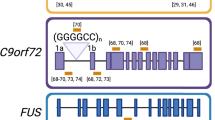
Several neurodegenerative diseases, including amyotrophic lateral sclerosis (ALS), have a complex genetic background, in addition to cases where the disease appears to manifest sporadically. The recent discovery of the hexanucleotide repeat expansion in the C9orf72 gene as the causative agent of ALS (C9ALS) gives rise to the opportunity to develop new therapies directed at this mutation , which is responsible for a large proportion of ALS and/or frontotemporal dementia cases. Mammalian models conscientiously replicating the late-onset motor defects and cellular pathologies seen in human patients do not exist. In this context, patient-derived cells give us a platform to test potential antisense oligonucleotide therapies, which could be the key to treat this subtype of motor neuron disease. Recently, we described that locked nucleic acid gapmer oligonucleotide-based treatment targeting C9orf72 repeat expanded transcripts resulted in recovery from the disease-related phenotypes in patient-derived fibroblasts. Our findings highlight the therapeutic potential of C9ALS using this gapmer oligonucleotide-based approach.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
DeJesus-Hernandez M, Mackenzie IR, Boeve BF et al (2011) Expanded GGGGCC Hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72:245–256
Renton AE, Majounie E, Waite A et al (2011) A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72:257–268
Gijselinck I, Van Langenhove T, van der Zee J et al (2012) A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol 11:54–65
Gendron TF, Bieniek KF, Zhang YJ et al (2013) Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol 126:829–844
Lagier-Tourenne C, Baughn M, Rigo F et al (2013) Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc Natl Acad Sci U S A 110(47):E4530–E4539
Cooper-Knock J, Higginbottom A, Stopford MJ et al (2015) Antisense RNA foci in the motor neurons of C9ORF72-ALS patients are associated with TDP-43 proteinopathy. Acta Neuropathol 130:63–75
van Blitterswijk M, Dejesus-hernandez M, Niemantsverdriet E et al (2013) Association between repeat sizes and clinical and pathological characteristics in carriers of C9ORF72 repeat expansions ( Xpansize-72 ): a cross-sectional cohort study. Lancet Neurol 12:978–988
van Blitterswijk M, Gendron TF, Baker MC et al (2015) Novel clinical associations with specific C9ORF72 transcripts in patients with repeat expansions in C9ORF72. Acta Neuropathol 130:863–876
Cruts M, Gijselinck I, Van Langenhove T et al (2013) Current insights into the C9orf72 repeat expansion diseases of the FTLD/ALS spectrum. Trends Neurosci 36:450–459
Almeida S, Gascon E, Tran H et al (2013) Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol 126:385–399
Haeusler AR, Donnelly CJ, Periz G et al (2014) C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature 507:195–200
Zhou B, Liu C, Geng Y et al (2015) Topology of a G-quadruplex DNA formed by C9orf72 hexanucleotide repeats associated with ALS and FTD. Sci Rep 5:16673
Sareen D, O’Rourke JG, Meera P et al (2013) Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci Transl med 5:208ra149. Sci Transl Med 5(208):208ra149
Cooper-Knock J, Walsh MJ, Higginbottom A et al (2014) Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions. Brain 137(Pt 7):2040–2051
Lee YB, Chen HJ, Peres J et al (2013) Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep 5:1178–1186
Mann DM, Robinson A, Rollinson S et al (2013) Dipeptide repeat proteins are present in the p62 positive inclusions in patients with frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9ORF72. Acta Neuropathol Commun 1:1
Mori K, Weng S, Arzberger T et al (2013) The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339:1335–1339
Ash PEA, Bieniek KF, Gendron TF et al (2013) Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77:639–646
Jovičić A, Mertens J, Boeynaems S et al (2015) Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci 18:1226–1229
Zhang K, Donnelly CJ, Haeusler AR et al (2015) The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 525(7567):56–61
Freibaum BD, Lu Y, Lopez-Gonzalez R et al (2015) GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature 525(7567):129–133
Moens TG, Niccoli T, Wilson KM et al (2019) C9orf72 arginine-rich dipeptide proteins interact with ribosomal proteins in vivo to induce a toxic translational arrest that is rescued by eIF1A. Acta Neuropathol 137(3):487–500
Donnelly CJ, Zhang PW, Pham JT et al (2013) RNA toxicity from the ALS / FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron 80:415–428
O’Rourke JG, Bogdanik L, Muhammad AKMG et al (2015) C9orf72 BAC transgenic mice display typical pathologic features of ALS/FTD. Neuron 88:892–901
Jiang J, Zhu Q, Gendron TF et al (2016) Gain of toxicity from ALS/FTD-linked repeat expansions in C9ORF72 is alleviated by antisense oligonucleotides targeting GGGGCC-containing RNAs. Neuron 90:535–550
Mendell JR, Al-Zaidy S, Shell R et al (2017) Single-dose gene-replacement therapy for spinal muscular atrophy. N Engl J Med 377:1713–1722
Shimizu-Motohashi Y, Miyatake S, Komaki H et al (2016) Recent advances in innovative therapeutic approaches for Duchenne muscular dystrophy: from discovery to clinical trials. Am J Transl Res 8:2471–2489
Levine TP, Daniels RD, Gatta AT et al (2013) The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN Rab-GEFs. 29:499–503
Marat AL, Dokainish H, McPherson PS (2011) DENN domain proteins: Regulators of Rab GTPases. J Biol Chem 286:13791–13800
Farg MA, Sundaramoorthy V, Sultana JM et al (2014) C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum Mol Genet 23:3579–3595
Aoki Y, Manzano R, Lee Y, et al (2017) C9orf72 and RAB7L1 regulate vesicle trafficking in amyotrophic lateral sclerosis and frontotemporal dementia. 140:887–897
Corbier C, Sellier C (2017) C9ORF72 is a GDP/GTP exchange factor for Rab8 and Rab39 and regulates autophagy. Small GTPases 8:181–186
Webster CP, Smith EF, Bauer CS, et al (2016) The C9orf72 protein interacts with Rab1a and the ULK1 complex to regulate initiation of autophagy. EMBO J 1–21
Yang M, Liang C, Swaminathan K et al (2016) A C9ORF72/SMCR8-containing complex regulates ULK1 and plays a dual role in autophagy. Neurosci Lett 1–17
Amick J, Roczniak-Ferguson A, Ferguson SM (2016) C9orf72 binds SMCR8, localizes to lysosomes, and regulates mTORC1 signaling. Mol Biol Cell 27:3040–3051
Sullivan PM, Zhou X, Robins AM et al (2016) The ALS/FTLD associated protein C9orf72 associates with SMCR8 and WDR41 to regulate the autophagy-lysosome pathway. Acta Neuropathol Commun 4:51
Obika S, Nanbu D, Hari Y et al (1997) Synthesis of 2′-O,4′-C-methyleneuridine and -cytidine. Novel bicyclic nucleosides having a fixed c3-endo sugar puckering. Tetrahedron Lett 38:8735–8738
Inoue H, Hayase Y, Iwai S et al (1987) Sequence-dependent hydrolysis of RNA using modified oligonucleotide splints and RNase H. FEBS Lett 215:327–330
Kurreck J, Wyszko E, Gillen C et al (2002) Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res 30:1911–1918
Fedoroff OY, Salazar M, Reid BR (1993) Structure of a DNA : RNA hybrid duplex. J Mol Biol 233(3):509–523
Crooke ST (1999) Molecular mechanisms of action of antisense drugs. Biochim Biophys Acta 1489:31–43
Shen W, De Hoyos CL, Migawa MT et al (2019) Chemical modification of PS-ASO therapeutics reduces cellular protein-binding and improves the therapeutic index. Nat Biotechnol 37:640–650
Migawa MT, Shen W, Wan WB et al (2019) Site-specific replacement of phosphorothioate with alkyl phosphonate linkages enhances the therapeutic profile of gapmer ASOs by modulating interactions with cellular proteins. Nucleic Acids Res 47:5465–5479
Acknowledgments
This work was supported by a Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research (C) [grant number 18 K11067 to N.M and 18 K07544 to Y.A.], Grants-in-Aid for Research on Nervous and Mental Disorders [grant number 28-6 to Y.A.], and the Japan Agency for Medical Research and Development [grant numbers 19ek0109239h0003 and 19lm0203086h0001 to Y.A.].
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Sathyaprakash, C. et al. (2020). Development of LNA Gapmer Oligonucleotide-Based Therapy for ALS/FTD Caused by the C9orf72 Repeat Expansion. In: Yokota, T., Maruyama, R. (eds) Gapmers. Methods in Molecular Biology, vol 2176. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-0771-8_14
Download citation
DOI: https://doi.org/10.1007/978-1-0716-0771-8_14
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-0770-1
Online ISBN: 978-1-0716-0771-8
eBook Packages: Springer Protocols