Abstract
Background
We developed models for tumor segmentation to automate the assessment of total tumor volume (TTV) in patients with colorectal liver metastases (CRLM).
Methods
In this prospective cohort study, pre- and post-systemic treatment computed tomography (CT) scans of 259 patients with initially unresectable CRLM of the CAIRO5 trial (NCT02162563) were included. In total, 595 CT scans comprising 8,959 CRLM were divided into training (73%), validation (6.5%), and test sets (21%). Deep learning models were trained with ground truth segmentations of the liver and CRLM. TTV was calculated based on the CRLM segmentations. An external validation cohort was included, comprising 72 preoperative CT scans of patients with 112 resectable CRLM. Image segmentation evaluation metrics and intraclass correlation coefficient (ICC) were calculated.
Results
In the test set (122 CT scans), the autosegmentation models showed a global Dice similarity coefficient (DSC) of 0.96 (liver) and 0.86 (CRLM). The corresponding median per-case DSC was 0.96 (interquartile range [IQR] 0.95–0.96) and 0.80 (IQR 0.67–0.87). For tumor segmentation, the intersection-over-union, precision, and recall were 0.75, 0.89, and 0.84, respectively. An excellent agreement was observed between the reference and automatically computed TTV for the test set (ICC 0.98) and external validation cohort (ICC 0.98). In the external validation, the global DSC was 0.82 and the median per-case DSC was 0.60 (IQR 0.29–0.76) for tumor segmentation.
Conclusions
Deep learning autosegmentation models were able to segment the liver and CRLM automatically and accurately in patients with initially unresectable CRLM, enabling automatic TTV assessment in such patients.
Relevance statement
Automatic segmentation enables the assessment of total tumor volume in patients with colorectal liver metastases, with a high potential of decreasing radiologist’s workload and increasing accuracy and consistency.
Key points
• Tumor response evaluation is time-consuming, manually performed, and ignores total tumor volume.
• Automatic models can accurately segment tumors in patients with colorectal liver metastases.
• Total tumor volume can be accurately calculated based on automatic segmentations.
Graphical Abstract

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Background
Response to systemic treatment of solid tumors is currently assessed using the Response Evaluation Criteria in Solid Tumors (RECIST1.1) [1, 2]. According to RECIST1.1, response to treatment is measured as the change in the sum of diameters in two target lesions per organ. RECIST1.1 aims to perform an objective assessment of tumor change, but the measurements are performed manually. This is not only tedious and time-consuming, but also subjective. The subjective nature of RECIST 1.1 leads to nonnegligible inter- and intra-observer variability [3, 4].
In patients with colorectal liver metastases (CRLM), the efficacy of RECIST1.1 has been questioned [5,6,7]. Colorectal cancer is the third most common cancer and the second leading cause of cancer-related deaths for men and women globally [8]. Almost half of these patients develop CRLM during the course of their illness [9,10,11]. For patients with CRLM, treatment response evaluation is crucial, as approximately 80% of these patients are not suitable for a potential curative local treatment at diagnosis [12, 13]. Patients with unresectable CRLM most often receive systemic treatment in a palliative setting or in a neoadjuvant setting to induce downsizing of the tumor load. Patients with initially unresectable liver-only CRLM can become eligible for local treatment with curative intent by systemic induction treatment in approximately 25% of cases [14,15,16].
Treatment decision-making for patients with CRLM is predominantly based on arguments involving technical resectability [17]. The question remains if local treatment such as surgery is clinically relevant for the individual patient. There is a growing interest in how a shift can be made from technically driven surgery to biologically driven surgery. Biologically driven surgery aims to select patients for the most optimal treatment to achieve long-term survival, taking into consideration tumor biology [18]. By doing so, the effects of systemic therapy could be underestimated by RECIST1.1, as it ignores potentially valuable information about total tumor volume (TTV). Assessment of TTV response to systemic therapy could represent a clinically more reliable evaluation since baseline TTV has shown to be prognostic for overall survival and change in TTV for recurrence-free survival in patients with CRLM, whereas RECIST1.1 has not [6, 7, 19].
In recent years, several studies demonstrated that volumetric assessment using algorithms increases the reproducibility of response assessments [6, 20,21,22]. In most of these studies, semiautomatic segmentation models are used to perform volumetric assessments [6, 7, 20,21,22]. Segmentation is the delineation of tissue structures on diagnostic imaging, resulting in 3D contours of these structures. The use of semiautomatic models, however, is still time-consuming and would be too labor-intensive to perform in daily practice. Fully automatic segmentation models could enable the automation of TTV evaluation.
Numerous autosegmentation models have been developed for the segmentation of livers and liver tumors on computed tomography (CT) or magnetic resonance imaging (MRI) [23]. Most studies on autosegmentation of liver tumors used imaging data from the Liver Tumor Segmentation Challenge (LiTS) [24,25,26,27]. The LiTS was conducted to compare state-of-the-art automated liver and tumor segmentation methods, and the dataset contained imaging data of various types of liver tumors [25]. For response monitoring of CRLM, it is far more important to optimize the performance for this disease, than for a wide range of tumors. Focusing on autosegmentation of CRLM, Vorontsov et al. developed a deep learning model with variable performance with Dice similarity coefficient (DSC) ranging from 0.14 to 0.68, depending on lesion size [27]. This model was trained and validated on CT scans of various liver tumors and tested on a small dataset of 26 CT scans comprising patients with CRLM. We hypothesize that with a larger and homogeneous population of patients suffering from CRLM only, the performance of deep learning-based tumor and liver segmentation can be improved.
In this study, we aim to develop deep learning models for automatic tumor segmentation of CRLM and the liver using a comprehensive training and test set of patients with initially unresectable CRLM. The secondary aim is to automate the assessment of TTV response to systemic therapy in such patients.
Methods
Development cohort
Study population
In this prospective cohort study, patients registered between November 2014 and April 2019 from the ongoing multicenter randomized clinical trial of the Dutch Colorectal Cancer Group, CAIRO5 (NCT02162563), were included for model development and testing [28]. The CAIRO5 trial aims to select the optimal systemic induction therapy for patients with initially unresectable liver-only CRLM (Additional file 1: S1). Patients are randomized between different systemic therapy combinations based on primary tumor site and genetic mutation status (RAS/BRAF). Treatment regimens consist of doublet or triplet chemotherapy in combination with targeted therapy. All included patients signed a written informed consent form, also allowing side studies such as the current one.
Imaging
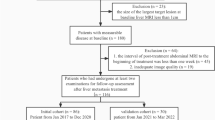
Imaging data of this development cohort consisted of contrast-enhanced CT scans of the chest and abdomen at baseline and every 2 months during systemic therapy. All scans were performed in one of the 54 medical centers responsible for inclusion using different types of CT scanners and acquisition protocols. In the current study, only patients with contrast-enhanced abdominal CT scans in the portal venous phase were included (Fig. 1). Exclusion criteria were non-contrast enhanced or not portal venous CT scan, missing or incomplete CT scan, the use of MRI or 18F-fluorodeoxyglucose positron emission tomography instead of portal venous CT, and technical error in segmentation software. CT acquisition characteristics are summarized in Additional file 1: S2.
Patient selection of development cohort. CT Computed tomography, MRI Magnetic resonance imaging. *The patients excluded because of “MRI scan” had a MRI scan instead of a CT scan for their diagnostic work-up. For patients with “Missing CT,” the baseline or follow-up CT scan was not available. The error in segmentation software occurred in the IntelliSpace Portal software of Philips
Data processing
Reference segmentations
All available pre- and post-treatment CT scans of the development cohort were used for semi-automatic segmentation of the liver and CRLM in the Tumor Tracking Modality of IntelliSpace Portal 9.0® (Philips Healthcare, Best, the Netherlands). In all CT scans, the liver and all CRLM were segmented by one of three trained members of the research team (N.J.W., S.P., R.K.). Lesions were roughly outlined, which resulted in a semi-automatic contour or region of interest based on differences in density. These contours were subsequently manually adjusted in every slice for accurate segmentation. All segmentations performed by the trained research team were verified and, if needed, adjusted by an abdominal radiologist with 18 years of experience (J.H.T.M.W.). Three abdominal radiologists with 10 (J.E.B.), 2 (I.M.N.), and 1 (S.I.M.) years of experience also independently corrected and verified 41 scans of 20 patients segmented by a member of the research team.
Image processing steps
The DICOM files of the CT scans and the DICOM-RT files of the 3D semi-automatic segmentations were uploaded into the SAS Viya® Analytical Platform (SAS Viya 3.5, SAS Institute Inc.). The scans and segmentations were combined to create liver and tumor masks which were used as target segmentation maps. The density values were adjusted by clipping and histogram equalization. Firstly, clipping between -100 and 400 Hounsfield units was performed to restrict the density values to a common range in the liver. Secondly, histogram equalization was applied to better distribute the image histogram, utilizing the full range of Hounsfield units in the histogram for every image evenly.
Development and testing of autosegmentation models
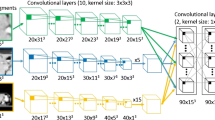
The U-net architecture was used for the segmentation models (Additional file 1: S3). Two U-nets were trained and tested, one for liver and one for tumor segmentation. Liver segmentation was performed to restrict the volume of interest for tumor segmentation. Model training, validation, and testing were performed within the SAS Viya® Analytical Platform. The radiologist’s segmentations of the liver and CRLM from the development cohort were used as reference data. A total of 434 (72.8%) CT scans with 6,667 CRLM were randomly assigned to the training set, 39 (6.5%) CT scans with 487 CRLM in the validation set, and 122 (20.6%) CT scans with 1,805 CRLM in the test set (Additional file 1: S4). The validation set was used for performance evaluation during training and to determine stop criteria. It was ensured that no image data of the same patient was included in both the training/validation set and the test set. This was done to prevent data leakage between the training/validation set and the test set. The automated liver segmentations were used as the volumes of interest for the autosegmentation tumor model (Fig. 2).
Automatic segmentation process. a The liver U-net model receives the computed tomography scan as input image. The output of the liver U-net model is the automatic liver segmentation. b The automatic liver segmentation is used as the volume of interest for the tumor U-net model. The output of the tumor U-net model is the automatic tumor segmentation
External validation
The tumor segmentation model performance was assessed with an external population of patients with CRLM. The CRLM dataset of the publicly available Workflow for Optimal Radiomics Classification, WORC, was used. This dataset consists of preoperative CT scans in the portal venous phase of 77 patients, surgically treated at the Erasmus University MC Rotterdam, the Netherlands (Additional file 1: S5) [29, 30]. All CRLM in the CT scans were segmented by one of the members of the research team and verified and if needed adjusted by an abdominal radiologist (J.H.T.M.W.) using IntelliSpace Portal 9.0® [31]. In addition, all livers and CRLM were automatically segmented by the developed models in the SAS Viya® Analytical Platform [32].
Statistics
The performances of the autosegmentation models and the segmentation agreement between different observers were assessed using the Dice similarity coefficient (DSC) as an accuracy measure, ranging between 0 (no overlap) and 1 (complete overlap) [33]. Two DSCs were calculated: the global DSC, which is the DSC of all CT scans combined, and the per-case DSC, which is the average per-CT scan DSC. Intersection-over-union, precision, and recall were also calculated. The summary statistics were calculated with formulas proposed by LiTS [25]. Total tumor volume was calculated in the SAS Viya® Analytical platform using the quantifyBioMedImages action [7, 34]. Total tumor volume was determined as the product of the voxel volume and the number of segmented voxels of all CRLM present in the liver and was reported as a continuous variable in cubic centimeters. A two-way mixed effect intraclass correlation coefficient (ICC) for absolute agreement was calculated to compare the reference and automatically computed TTV. The ICC was categorized as having either poor (ICC < 0.40), fair (ICC 0.40–0.59), good (ICC 0.60–0.74), or excellent (ICC 0.75–1.0) agreement [35, 36]. The distribution of normality of continuous variables was checked by visually inspecting the histograms and boxplots. Continuous variables were reported as median with interquartile range (IQR) and compared with Mann–Whitney U or t test, as appropriate. Categorical variables were displayed as frequencies and percentages and compared with chi-square test or Fisher’s exact test, as appropriate. Test results were considered statistically significant with a p < 0.05. Statistical analyses were performed using SAS® Studio (version 5.2, SAS Viya® 03.05).
Results
Study population
In total, 259 of 407 patients from the CAIRO5 trial were included in the development cohort of this study. The most common reason for exclusion was a missing CT scan, and 39 patients were not eligible because of not meeting inclusion criteria or withdrawal from the study (Fig. 1). Of all 259 patients, a baseline and first follow-up CT scan were available for analysis. In some cases, two or three follow-up scans were available and included. In total, 595 CT scans were included and 8,959 CRLM were segmented. In the development cohort, the median age was 62 (IQR 55−71) years and 36% (94/259) of the patients were female. Per patient, the median number of CRLM at baseline was 11 (IQR 7−21), with a median of six liver segments involved (IQR 4−7). Significant differences between training/validation and test set were observed, as a larger number of males were allocated in the training cohort, and the largest diameter of CRLM was smaller in the test set (Table 1). In the external validation cohort, a total of 72 patients with 112 CRLM were included. Five patients were excluded (Additional file 1: S5). The median age was 68 (IQR 59−77) years, 42% (30/72) of the patients were female, and the median number CRLM was 1 (IQR 1−2).
Accuracy of autosegmentation models
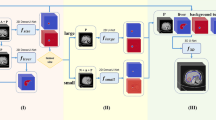
In the test set, the spatial agreement assessment of the autosegmentation models had a global DSC of 0.96 and 0.86 for liver and CRLM segmentation, respectively. The corresponding median per-case DSCs were 0.96 (IQR 0.95–0.96) and 0.80 (IQR 0.67–0.87). The intersection-over-union, precision, and recall were 0.75, 0.89, and 0.84 for tumor segmentation, respectively (Table 2). In Fig. 3, examples of the automatic segmentations of the liver and CRLM in the development cohort are depicted. Figure 4 illustrates a 3D visualization of automated liver and CRLM segmentations for three patients. The external validation cohort contained 72 CT scans. The autosegmentation tumor model resulted in a global DSC of 0.82 for CRLM segmentation, with a corresponding median per-case DSC of 0.60 (IQR 0.27–0.76). The intersection-over-union, precision, and recall were 0.69, 0.85, and 0.78 for tumor segmentation, respectively (Table 2). Figure 5 shows examples of the CRLM segmentation in two patients of the external validation.
Automatic segmentations of the liver and colorectal liver metastases in three patients of the development cohort. a, d, g Computed tomography scans before automatic segmentation. b, e, h Radiologist’s liver segmentation (pink) and automatic liver segmentation (blue). c, f, i Radiologist’s tumor segmentation (dark green) and automatic tumor segmentation (red)
Total tumor volume assessment
An excellent agreement was found between reference and automated TTV in the test set of the development cohort (ICC 0.97, confidence interval 95% 0.96–0.98) and in the external validation cohort (ICC 0.98, confidence interval 95% 0.96–0.99). In the development cohort, no significant difference (p = 0.632) was found in the reference TTV between the training cohort and the test set (Table 3).
Agreement between different observers
An excellent agreement in segmentation was found between the four independent expert abdominal radiologists in 41 scans of 20 patients. The per-case DSC ranged between 0.90 and 0.94 and the global DSC ranged between 0.91 and 0.94. The per-case DSC between the radiologist determining the ground truth and the three independent expert abdominal radiologists was 0.90, 0.92, and 0.91 (Table 4). In addition, a median per-case DSC of 0.99 was observed between the segmentations of the research team and the expert radiologist determining the ground truth.
Discussion
In this study, deep learning models were successfully developed to segment the liver and CRLM automatically and accurately in CT scans of patients suffering from initially unresectable CRLM. Moreover, the models enabled automatic assessment of TTV of all the CRLM in those CT scans with an excellent agreement with the radiologist’s assessment. In the external validation cohort, consisting of patients with upfront resectable CRLM, the models performed less accurately than in the test set of the development cohort.
The performances of the autosegmentation models in the CAIRO5 test set of this study were comparable or superior to autosegmentation models for liver and liver tumor segmentation in earlier studies [24,25,26]. In the LiTS, the best liver segmentation model scored a per-case DSC of 0.97, and the best tumor segmentation model scored a per-case DSC of 0.83 [25]. In contrast to this study, the LiTS Benchmark dataset contained imaging data of patients with different types of liver tumors.
The autosegmentation tumor model in the current study obtained lower DSCs in the external validation cohort. This could be explained by the different types of patients in the two data sets. The autosegmentation models were trained and tested on data consisting of pre- and post-treatment CT scans of patients with initially unresectable CRLM [28]. This patient group was initially not suitable for local therapy because of disease extensiveness and the liver CT scans were often complicated by confluent tumors and extensive numbers of CRLM. As a result, patients with a small number of metastases were underrepresented. We hypothesized that the autosegmentation tumor model capable of segmenting patients with extensive CRLM would also be capable of segmenting patients with less extensive disease.
The median smaller size of CRLM included in the external validation cohort could also be a reason for the lower DSCs. This was also demonstrated in the study of Vorontsov et al. [27], who developed deep learning models with the same U-net-architecture for automatic segmentation of CRLM in CT scans. In the test set of their study, the automatic model performed better in lesions larger than 20 mm as compared to lesions smaller than 10 mm or between 10 and 20 mm, obtaining per-lesion DSCs of 0.68, 0.14, and 0.53, respectively.
Autosegmentation remains a challenging task due to variable image parameters, patient variability, and tumor morphology. Therefore, autosegmentation models should be trained on CT scan data that is as realistic and robust as possible. In the current study, the CT acquisition parameters varied considerably across the 54 centers in the development cohort, since scans were performed using different CT scanners and acquisition protocols. However, all scans were of adequate quality to be used for patient management. The variety in CT acquisition parameters is a good representation of CT scans in daily practice and could be considered as a strength with respect to external validity.
The autosegmentation models allowed for the automatic assessment of TTV, not only leading to a more advanced interpretation of change in tumor size, as the effect on all tumorous tissue of all metastases is taken into account. In addition, this method is potentially also less subjective, tedious, and time-consuming than tumor response assessments by radiologists in the future. Assessment of TTV response to systemic therapy could represent a clinically more reliable tumor evaluation than RECIST1.1, as it was shown to be prognostic for recurrence-free survival, whilst RECIST1.1 was not [7]. Moreover, the autosegmentation models can enable the automatic assessment of other relevant imaging features for tumor response evaluation, such as morphological changes [5, 37]. Besides improving tumor response evaluation, the autosegmentation models could also play a role in radiomics research. Tumor segmentation forms an important step in the process of radiomics, in which hundreds of imaging features can be analyzed out of tumor segmentations and used in predictive modeling through machine learning [38,39,40].
It is important to emphasize that the autosegmentation models in the current study have been developed to improve tumor response evaluation of CRLM and not to diagnose CRLM. Models capable of diagnosing CRLM require a different approach with an extensive amount of data comprising different benign and malignant types of liver lesions.
During the design of the current study, the U-net was the state-of-the-art architecture, and a 2D U-net was employed instead of a 3D U-net. Recently, other architectures like the U-net + + and Trans U-net were developed, so it could be considered to make use of such architectures in the future. Moreover, the 2D U-Net was preferred over the 3D U-Net as it is more accurate specifically for the liver and requires less computational power [41].
The present study has several limitations. Firstly, the ground truth was based on the evaluation and adjustment of one expert radiologist. Consequently, the ground truth of one observer had a large influence on model training and ultimately model performance. The original study of the external CRLM cohort already reported significant differences between the segmentations of different observers [30]. However, excellent agreement in tumor segmentation was observed between four independent expert radiologists and it was not logistically feasible to base the ground truth on the segmentations of multiple radiologists. Therefore, it was chosen to determine the ground truth based on one radiologist. Secondly, a selection of patients with initially unresectable CRLM was used for model training. This may have influenced the generalizability of the developed autosegmentation tumor model, as it performed less in the external cohort consisting of patients with resectable and fewer number of CRLM. However, the autosegmentation models are developed to improve the evaluation of CRLM to systemic therapy. Patients with CRLM receiving systemic treatment often have more extensive disease or large tumors. Finally, to enhance density differences between the liver and tumors we have applied histogram equalization. However, this approach may have reduced the (calibrated) intensity values in the images for the segmentation steps. With the high accuracies obtained in our study, we do not expect that this pre-processing step has negatively influenced the segmentation agreement.
In the future, the actual implementation of an automatic tumor response pipeline into clinical care will face challenges such as technical feasibility, ethical concerns, and regulatory aspects [42, 43]. A potential first step to implementation is to conduct a prospective clinical study with an integrated tumor response pipeline with a human-in-the-loop situation. Moreover, if the automatic tumor response pipeline is implemented successfully and has proven to be clinically relevant, the autosegmentation model could be translated to other imaging modalities (e.g., MRI).
In conclusion, the deep learning models developed in this study were able to automatically segment the liver and CRLM with high accuracy in patients with initially unresectable CRLM. This has a high potential of decreasing radiologist’s workload and increasing accuracy by lowering interobserver variability. Moreover, the models enabled automatic assessment of TTV and the response of TTV to systemic treatment. This and other potentially highly relevant imaging features, such as tumor morphological response could potentially contribute to more consistent and clinically relevant tumor response assessments for patients with CRLM receiving systemic treatment in future clinical care and research.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- CRLM:
-
Colorectal liver metastases
- CT:
-
Computed tomography
- DSC:
-
Dice similarity coefficient
- ICC:
-
Intraclass correlation coefficient
- IQR:
-
Interquartile range
- LiTS:
-
Liver Tumor Segmentation Challenge
- MRI:
-
Magnetic resonance imaging
- TTV:
-
Total tumor volume
References
Therasse P, Arbuck SG, Eisenhauer EA et al (2000) New guidelines to evaluate the response to treatment in solid tumors. European organization for research and treatment of cancer, national cancer institute of the United States, national cancer institute of Canada. J Natl Cancer Inst 92:205–216. https://doi.org/10.1093/jnci/92.3.205
Eisenhauer EA, Therasse P, Bogaerts J et al (2009) New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 45:228–247. https://doi.org/10.1016/j.ejca.2008.10.026
Yoon SH, Kim KW, Goo JM, Kim DW, Hahn S (2016) Observer variability in RECIST-based tumour burden measurements: a meta-analysis. Eur J Cancer 53:5–15. https://doi.org/10.1016/j.ejca.2015.10.014
Beaumont H, Evans TL, Klifa C et al (2018) Discrepancies of assessments in a RECIST 11 phase I.I clinical trial - association between adjudication rate and variability in images and tumors selection. Cancer Imaging 18:50. https://doi.org/10.1186/s40644-018-0186-0
Chun YS, Vauthey JN, Boonsirikamchai P et al (2009) Association of computed tomography morphologic criteria with pathologic response and survival in patients treated with bevacizumab for colorectal liver metastases. JAMA 302:2338–2344. https://doi.org/10.1001/jama.2009.1755
Rothe JH, Grieser C, Lehmkuhl L et al (2013) Size determination and response assessment of liver metastases with computed tomography–comparison of RECIST and volumetric algorithms. Eur J Radiol 82:1831–1839. https://doi.org/10.1016/j.ejrad.2012.05.018
Wesdorp NJ, Bolhuis K, Roor J et al (2021) The prognostic value of total tumor volume response compared with RECIST1.1 in patients with initially unresectable colorectal liver metastases undergoing systemic treatment. Ann Surg Open. 2:e103. https://doi.org/10.1097/as9.0000000000000103
Sung H, Ferlay J, Siegel RL et al (2021) Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 71:209–249. https://doi.org/10.3322/caac.21660
Elferink MAG, de Jong KP, Klaase JM, Siemerink EJ, de Wilt JHW (2015) Metachronous metastases from colorectal cancer: a population-based study in North-East Netherlands. Int J Colorectal Dis 30:205–212. https://doi.org/10.1007/s00384-014-2085-6
van der Geest LGM, Jt L-B, Koopman M et al (2015) Nationwide trends in incidence, treatment and survival of colorectal cancer patients with synchronous metastases. Clin Exp Metas 32:457–465. https://doi.org/10.1007/s10585-015-9719-0
Van Cutsem E, Cervantes A, Adam R et al (2016) ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann Oncol 27:1386–1422. https://doi.org/10.1093/annonc/mdw235
de Ridder JAM, van der Stok EP, Mekenkamp LJ et al (2016) Management of liver metastases in colorectal cancer patients: a retrospective case-control study of systemic therapy versus liver resection. Eur J Cancer 59:13–21. https://doi.org/10.1016/j.ejca.2016.02.003
Noren A, Eriksson HG, Olsson LI (2016) Selection for surgery and survival of synchronous colorectal liver metastases; a nationwide study. Eur J Cancer 53:105–114. https://doi.org/10.1016/j.ejca.2015.10.055
Nordlinger B, Van Cutsem E, Rougier P et al (2007) Does chemotherapy prior to liver resection increase the potential for cure in patients with metastatic colorectal cancer? A report from the European Colorectal Metastases Treatment Group. Eur J Cancer 43:2037–2045. https://doi.org/10.1016/j.ejca.2007.07.017
Adam R, Kitano Y (2019) Multidisciplinary approach of liver metastases from colorectal cancer. Ann Gastroenterol Surg 3:50–56. https://doi.org/10.1002/ags3.12227
Lam VW, Spiro C, Laurence JM et al (2012) A systematic review of clinical response and survival outcomes of downsizing systemic chemotherapy and rescue liver surgery in patients with initially unresectable colorectal liver metastases. Ann Surg Oncol 19:1292–1301. https://doi.org/10.1245/s10434-011-2061-0
Adams RB, Aloia TA, Loyer E et al (2013) Selection for hepatic resection of colorectal liver metastases: expert consensus statement. HPB (Oxford) 15:91–103. https://doi.org/10.1111/j.1477-2574.2012.00557.x
Bolhuis K, Kos M, van Oijen MGH, Swijnenburg RJ, Punt CJA (2020) Conversion strategies with chemotherapy plus targeted agents for colorectal cancer liver-only metastases: a systematic review. Eur J Cancer 141:225–238. https://doi.org/10.1016/j.ejca.2020.09.037
Tai K, Komatsu S, Sofue K et al (2020) Total tumour volume as a prognostic factor in patients with resectable colorectal cancer liver metastases. BJS Open. https://doi.org/10.1002/bjs5.50280
van Kessel CS, van Leeuwen MS, Witteveen PO et al (2012) Semi-automatic software increases CT measurement accuracy but not response classification of colorectal liver metastases after chemotherapy. Eur J Radiol 81:2543–2549. https://doi.org/10.1016/j.ejrad.2011.12.026
Lin M, Pellerin O, Bhagat N et al (2012) Quantitative and volumetric European Association for the Study of the Liver and Response Evaluation Criteria in Solid Tumors measurements: feasibility of a semiautomated software method to assess tumor response after transcatheter arterial chemoembolization. J Vasc Interv Radiol 23:1629–1637. https://doi.org/10.1016/j.jvir.2012.08.028
Yan J, Schwartz LH, Zhao B (2015) Semiautomatic segmentation of liver metastases on volumetric CT images. Med Phys 42:6283–6293. https://doi.org/10.1118/1.4932365
Chu LC, Park S, Kawamoto S et al (2021) Current status of radiomics and deep learning in liver imaging. J Comput Assist Tomogr 45:343–351. https://doi.org/10.1097/rct.0000000000001169
Chlebus G, Schenk A, Moltz JH et al (2018) Automatic liver tumor segmentation in CT with fully convolutional neural networks and object-based postprocessing. Sci Rep 8:15497. https://doi.org/10.1038/s41598-018-33860-7
Bilic P, Christ P, Li HB et al (2019). The Liver Tumor Segmentation Benchmark (LiTS). arXiv preprint arXiv:190104056.
Vorontsov E, Chartrand G, Tang A, Pal C, Kadoury S (2018). Liver lesion segmentation informed by joint liver segmentation. 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018).1332–5.
Vorontsov E, Cerny M, Régnier P et al (2019) Deep learning for automated segmentation of liver lesions at CT in patients with colorectal cancer liver metastases. Radiology 1:180014. https://doi.org/10.1148/ryai.2019180014
Huiskens J, van Gulik TM, van Lienden KP et al (2015) Treatment strategies in colorectal cancer patients with initially unresectable liver-only metastases, a study protocol of the randomised phase 3 CAIRO5 study of the Dutch Colorectal Cancer Group (DCCG). BMC Cancer 15:365. https://doi.org/10.1186/s12885-015-1323-9
Starmans MPA, Timbergen MJM, Vos M, et al. (2021). The WORC database: MRI and CT scans, segmentations, and clinical labels for 930 patients from six radiomics studies. medRxiv.2021.08.19.21262238. https://doi.org/10.1101/2021.08.19.21262238.
Starmans MPA, Buisman FE, Renckens M et al (2021) Distinguishing pure histopathological growth patterns of colorectal liver metastases on CT using deep learning and radiomics: a pilot study. Clin Exp Metas 38:483–494. https://doi.org/10.1007/s10585-021-10119-6
Philips: IntelliSpace Portal 9.0: Advanced visual analysis. https://www.usa.philips.com/healthcare/product/HC881072/intellispace-portal-advanced-visualization-solution Accessed 2023.
SAS: SAS visual analytics. https://www.sas.com/en_us/software/visual-analytics.html Accessed 2023.
Zijdenbos AP, Dawant BM, Margolin RA, Palmer AC (1994) Morphometric analysis of white matter lesions in MR images: method and validation. IEEE Trans Med Imaging 13:716–724. https://doi.org/10.1109/42.363096
SAS Visual Data Mining and Machine Learning Programming Guide: The quantifyBioMedImages Action. https://go.documentation.sas.com/?cdcId=pgmsascdc&cdcVersion=9.4_3.5&docsetId=casactml&docsetTarget=casactml_biomedimage_details05.htm&locale=en (2020). Accessed 28–12–2020 2020.
Cicchetti D (1994) Guidelines, criteria, and rules of thumb for evaluating normed and standardized assessment instruments in psychology. Psychol Assess 6:284–290
Hallgren KA (2012) Computing inter-rater reliability for observational data: an overview and tutorial. Tutor Quant Methods Psychol 8:23–34. https://doi.org/10.20982/tqmp.08.1.p023
Shindoh J, Loyer EM, Kopetz S et al (2012) Optimal morphologic response to preoperative chemotherapy: an alternate outcome end point before resection of hepatic colorectal metastases. J Clin Oncol 30:4566–4572. https://doi.org/10.1200/jco.2012.45.2854
Lambin P, Leijenaar RTH, Deist TM et al (2017) Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 14:749–762. https://doi.org/10.1038/nrclinonc.2017.141
Lambin P, Rios-Velazquez E, Leijenaar R et al (2012) Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 48:441–446. https://doi.org/10.1016/j.ejca.2011.11.036
Wesdorp NJ, Hellingman T, Jansma EP et al (2020) Advanced analytics and artificial intelligence in gastrointestinal cancer: a systematic review of radiomics predicting response to treatment. Eur J Nucl Med Mol Imaging. https://doi.org/10.1007/s00259-020-05142-w
Zettler N, Mastmeyer A (2021) Comparison of 2D vs. 3D U-Net Organ Segmentation in abdominal 3D CT images. arXiv. https://doi.org/10.48550/arXiv.2107.04062
van de Sande D, van Genderen ME, Huiskens J, Gommers D, van Bommel J (2021) Moving from bytes to bedside: a systematic review on the use of artificial intelligence in the intensive care unit. Intensive Care Med 47:750–760. https://doi.org/10.1007/s00134-021-06446-7
van de Sande D, Van Genderen ME, Smit JM et al (2022) Developing, implementing and governing artificial intelligence in medicine: a step-by-step approach to prevent an artificial intelligence winter. BMJ Health Care Inform 29:e100495. https://doi.org/10.1136/bmjhci-2021-100495
Acknowledgements
We thank all patients who consented to participate in the CAIRO5 study. We would like to acknowledge support from the Dutch Cancer Society (KWF Kankerbestrijding) and unrestricted grants from the Cancer Center Amsterdam Foundation, Roche, and Amgen. SAS supported the study in kind by providing advanced analytics and deep learning expertise.
Funding
The CAIRO5 study is supported by unrestricted scientific grants from Roche and Amgen. The current study is supported by the Dutch Cancer Society (KWF Kankerbestrijding), project number 14002/2021–2, and an unrestricted grant from the Cancer Center Amsterdam Foundation. SAS supported the study in kind by providing advanced analytics and deep learning expertise. Funders had no role in the design, conduct, analysis of the study, writing of the manuscript, nor in the decision to submit the manuscript for publication.
Author information
Authors and Affiliations
Contributions
Research design: NJW, JH, and GK. Manuscript writing: NJW, JMZ, HAM, RJS, JS, CJAP, JH, and GK. Performance of the research: NJW, JMZ, JR, JHTMW, RK, FV, CA, SD, MJA, TC, CHCD, MRWE, MFG, DG, TMG, JJH, KPJ, JMK, MSLL, KPL, IQM, GAP, AMR, TMR, CV, JHHW, RJS, CJAP, and GK. Data collection: NJW, SP, JR, RK, JHTMW, JVDB, IN, and SM. Data analysis: NJW, JMZ, JR, SD, HAM, JS, RJS, CJAP, JH, and GK. Critical revision of the manuscript: All authors.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The study was conducted according to the ethical standards of the Helsinki Declaration of 1975 and has been approved by the medical ethical committee (METc VUmc; 23–04-2019 (2019.236)). All included patients signed a written informed consent form, also allowing side studies such as the current one.
Consent for publication
Not applicable.
Competing interests
CJAP has an advisory role for Nordic Pharma. HAM is a co-founder and shareholder of Nicolab.
JS is a member of the European Radiology Experimental Editorial Board. He has not taken part in the review or selection process of this article.
All remaining authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
S1. Inclusion and exclusion criteria of the CAIRO5 trial[1]. S2. CT parameters. S3. Model details. S4. Scans in training, validation and test set. S5. Flow diagram patient selection external validation cohort.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wesdorp, N.J., Zeeuw, J.M., Postma, S.C.J. et al. Deep learning models for automatic tumor segmentation and total tumor volume assessment in patients with colorectal liver metastases. Eur Radiol Exp 7, 75 (2023). https://doi.org/10.1186/s41747-023-00383-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41747-023-00383-4