Abstract
Due to advances in genomic technologies, our understanding of estrogen receptor (ER)-mediated transcription in breast cancer cells has evolved significantly in recent years. Genome-wide mapping experiments revealed thousands of ER-binding events, but linking them to the target genes has been an ongoing struggle. A recent paper describes a new technique, called ChIA-PET (chromatin interaction analysis using paired-end tag sequencing), that can directly address these questions. ChIA-PET is an unbiased approach for simultaneously identifying all genome-wide binding events of a transcription factor and those involved in long-range chromatin loops.
Similar content being viewed by others
Background
Estrogen receptor (ER) has been one of the most intensively studied transcription factors, primarily due to its importance in breast cancer. Much effort has been invested in identifying the target genes of ER, mainly by expression arrays in breast cancer cell lines and in human tumour samples [1, 2]. In the past few years, the evolution of genomic technologies has permitted mapping of ER-binding events in an unbiased manner [3, 4]. This involves combining chromatin immunoprecipitation with microarrays or high-throughput sequencing in order to identify ER contact points within the genome. These genome-wide experiments revealed a number of surprising features about ER biology, the most unexpected one being that ER regulates transcription mostly from distal enhancers. This finding raised the subsequent question: which of the thousands of ER-binding events are the functional ones that regulate expression of the few hundred ER target genes? Fullwood and colleagues [5] have now established a method for addressing this question, allowing researchers to assign ER-binding events to transcriptional hubs and target genes.
The article
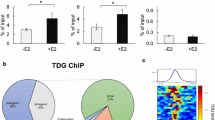
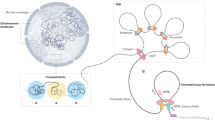
Researchers at the Genome Institute of Singapore have developed a technique called ChIA-PET (chromatin interaction analysis using paired-end tag sequencing) [5], which couples chromosome conformation capture [6], a method for identifying interacting chromatin regions, with high-throughput sequencing. This permits the global identification of chromatin loops formed at a specific time point. The authors used a well-established system whereby MCF-7 breast cancer cells are cultured in hormone-deprived media and subsequently treated with estrogen for a short time period, resulting in synchronisation of transcriptional events [7]. Using this system, the authors could capture the high-confidence chromatin interactions that form during the first wave of estrogen-ER-mediated transcription. These interactions were extensively validated, supporting their conclusions. The implications of this investigation are profound. It is revealed that chromatin interactions can occur in clusters of contact points and that the actual anchor points, in a linear two-dimensional representation, can be found separated by hundreds of kilobases and in some cases megabases. Contrary to previous suggestions [8], inter-chromosomal interactions do not appear to be a common or experimentally verifiable phenomenon. This suggests that ER-binding events regulate genes within mostly a spatially confined environment, which in many cases involves the folding of multiple regulatory regions into one transcriptional hub.
In summary, the authors find 689 ER-associated chromatin interaction complexes made up of duplexes and more complex interactions. These tend to involve the stronger ER-binding events, which are biased toward specific histone marks and other transcriptional regulators important for ER function, such as FoxA1 [9]. By combining the ChIA-PET data with gene expression data, they could discover individual transcriptional hubs and the target genes localised in the vicinity.
The viewpoint
Until now, researchers have been linking enhancers and cis-regulatory elements with the target genes one at a time [10, 11]. ChIA-PET circumvents this limitation and can identify all active loops at a given time point, allowing us to redefine the functional ER-binding events. While this constitutes the first snapshot of ER-mediated chromatin loops, it is a quantum leap in our understanding of transcription dynamics. However, this pioneering work leads to a series of new questions. The authors associate the DNA loops that they captured at 45 minutes of estrogen treatment with a time course expression profile of cells treated with estrogen from 0 to 48 hours of treatment. A challenging concept that needs further testing is whether the chromatin loops (captured at 45 minutes) are stable over time or whether they change, leading to the formation of new loops at later time points to regulate transcription of the 'late' target genes. Moreover, what is the role of co-operating transcription factors in loop formation? Also, since many of the ER functional binding events do not directly involve promoter regions, one can speculate that some DNA loops might circum vent the need for a transcription factor to be present at the promoter by concentrating the transcription machinery in the three-dimensional hubs.
Furthermore, this technology can be used to explore ER chromatin loops and transcriptional hubs in different breast cancer cell lines and treatment conditions. The identification of transcriptionally active binding events in the different contexts will illuminate the factors that dictate differential binding and transcriptional activity. However, the major limitations of the ChIA-PET technology are the complex technical aspects of the procedure, the heavy bioinformatics demand and the requirement for high-throughput sequencers. Nevertheless, these genomic tools are becoming more readily available, and as such, ChIA-PET will prove to be a powerful tool for delineating mechanisms of transcription in a spatial and temporal manner, applicable to any transcription factor of interest.
Abbreviations
- ChIA-PET:
-
chromatin interaction analysis using paired-end tag sequencing
- ER:
-
estrogen receptor.
References
Perou CM, Sørlie T, Eisen MB, Rijn van de M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lønning PE, Børresen-Dale AL, Brown PO, Botstein D: Molecular portraits of human breast tumours. Nature. 2000, 406: 747-752. 10.1038/35021093.
Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW: A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006, 10: 515-527. 10.1016/j.ccr.2006.10.008.
Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M: Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006, 38: 1289-1297. 10.1038/ng1901.
Lin CY, Vega VB, Thomsen JS, Zhang T, Kong SL, Xie M, Chiu KP, Lipovich L, Barnett DH, Stossi F, Yeo A, George J, Kuznetsov VA, Lee YK, Charn TH, Palanisamy N, Miller LD, Cheung E, Katzenellenbogen BS, Ruan Y, Bourque G, Wei CL, Liu ET: Whole-genome cartography of estrogen receptor alpha binding sites. PLoS Genet. 2007, 3: e87-10.1371/journal.pgen.0030087.
Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, Chew EG, Huang PY, Welboren WJ, Han Y, Ooi HS, Ariyaratne PN, Vega VB, Luo Y, Tan PY, Choy PY, Wansa KD, Zhao B, Lim KS, Leow SC, Yow JS, Joseph R, Li H, Desai KV, Thomsen JS, Lee YK, et al: An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009, 462: 58-64. 10.1038/nature08497.
Dekker J, Rippe K, Dekker M, Kleckner N: Capturing chromosome conformation. Science. 2002, 295: 1306-1311. 10.1126/science.1067799.
Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M: Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000, 103: 843-852. 10.1016/S0092-8674(00)00188-4.
Hu Q, Kwon YS, Nunez E, Cardamone MD, Hutt KR, Ohgi KA, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG, Fu XD: Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci USA. 2008, 105: 19199-19204. 10.1073/pnas.0810634105.
Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M: FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008, 132: 958-970. 10.1016/j.cell.2008.01.018.
Barnett DH, Sheng S, Charn TH, Waheed A, Sly WS, Lin CY, Liu ET, Katzenellenbogen BS: Estrogen receptor regulation of carbonic anhydrase XII through a distal enhancer in breast cancer. Cancer Res. 2008, 68: 3505-3515. 10.1158/0008-5472.CAN-07-6151.
Pan YF, Wansa KD, Liu MH, Zhao B, Hong SZ, Tan PY, Lim KS, Bourque G, Liu ET, Cheung E: Regulation of estrogen receptor-mediated long range transcription via evolutionarily conserved distal response elements. J Biol Chem. 2008, 283: 32977-32988. 10.1074/jbc.M802024200.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Rights and permissions
About this article
Cite this article
Theodorou, V., Carroll, J.S. Estrogen receptor action in three dimensions - looping the loop. Breast Cancer Res 12, 303 (2010). https://doi.org/10.1186/bcr2470
Published:
DOI: https://doi.org/10.1186/bcr2470