Abstract
Background
The glycolytic pathway is central to cellular energy production. Selection on individual enzymes within glycolysis, particularly phosphoglucose isomerase (Pgi), has been associated with metabolic performance in numerous organisms. Nonetheless, how whole energy-producing pathways evolve to allow organisms to thrive in different environments and adopt new lifestyles remains little explored. The Lanceocercata radiation of Australasian stick insects includes transitions from tropical to temperate climates, lowland to alpine habitats, and winged to wingless forms. This permits a broad investigation to determine which steps within glycolysis and what sites within enzymes are the targets of positive selection. To address these questions we obtained transcript sequences from seven core glycolysis enzymes, including two Pgi paralogues, from 29 Lanceocercata species.
Results
Using maximum likelihood methods a signature of positive selection was inferred in two core glycolysis enzymes. Pgi and Glyceraldehyde 3-phosphate dehydrogenase (Gaphd) genes both encode enzymes linking glycolysis to the pentose phosphate pathway. Positive selection among Pgi paralogues and orthologues predominately targets amino acids with residues exposed to the protein’s surface, where changes in physical properties may alter enzyme performance.
Conclusion
Our results suggest that, for Lancerocercata stick insects, adaptation to new stressful lifestyles requires a balance between maintaining cellular energy production, efficiently exploiting different energy storage pools and compensating for stress-induced oxidative damage.
Similar content being viewed by others
Background
Respiration generates the cellular energy required for the fundamental biological processes of growth, development and reproduction. The pathways involved in respiration are well characterised, flowing from glycolysis to the Krebs cycle, along with well-understood branch points, including the pentose phosphate pathway activated during periods of oxidative stress [1] and increased lipid biosynthesis [2]. Nonetheless, how these pathways as a whole evolve to allow organisms to live in different environments and adopt new life history traits that might require altered energy levels requires further attention [3].
Enzymes in metabolic pathways are kinetically entwined in a complex conveyer belt of substrate to end-product [4]. As a result of this interdependence, mutations that alter in vitro catalytic activity may not alter the observed in vivo phenotype [4]. Enzymes with the greatest flux control through pathways are likely targets of natural selection [5–7], due to the probability of fixation of any adaptive change associated to its conferred advantage. Flux control may be proportionally distributed across the pathway [4] or asymmetrically localised in enzymes at the top of the pathway [8, 9]. The evolution and incidence of selection is dependent on the properties and complexities of the pathway, with the disparity between current and optimal flux being a key factor [9]. The evolution of flux control and selection on enzyme polymorphisms has been intensively studied in the glycolytic pathway of Drosophila[3, 6, 10, 11].
Glycolysis comprises a series of ten enzyme-catalysed reactions that convert glucose to pyruvate, the substrate for the Krebs cycle. Concurrently, glycolysis also generates small quantities of ATP and NADH that are utilised by downstream high-energy yielding processes. Although glycolysis is an essential biological pathway conserved across all eukaryotes, the component enzymes exhibit extensive variation at both the nucleotide and protein levels, which may be the target of selection [3]. In Drosophila, positive selection is centred on the glucose-6-phosphate intersection for glycolysis and the pentose phosphate pathway [11]. The branch point enzymes are hypothesised to possess novel flux control resulting in higher selection coefficients for adaptive mutations [3]. In the Drosophila species studied, branch point enzymes accumulate more adaptive mutations than enzymes at the top of the pathway, which are conserved by strong purifying selection [10]. Simulations provide theoretical support for this empirical evidence, and make the prediction that the further flux is from optimum, the more likely adaptive substitutions are to occur disproportionately in branching enzymes [9]. Further work is required in species with diverse metabolic demands to test the generality of the Drosophila findings [3]. Positive selection and adaptation within glycolysis and its connecting pathways are likely to be a balance between maintaining cellular energy production, reducing the associated damage caused by oxidative stress [1], and efficiently exploiting the available substrate storage pools (e.g. glycogen or triglycerides) [6].
A majority of evidence for selection within the glycolytic enzymes, and its correlation with fitness, has been accumulated for the dimeric enzyme phosphoglucose isomerase (PGI; EC 5.3.1.9), which is not a rate-limiting step in glycolysis [10]. Variation in Pgi has been associated with flight performance, thermal adaptation and other fitness-related traits in a diverse array of organisms [12, 13], including arthropods [14–18], bacteria [19], molluscs [20], and plants [21]. PGI is thought to be under climate-related directional selection in the beetle Chrysomela aeneicollis (Coleoptera), where distinct alleles with functional and physiological differences are restricted to cold or warm habitats [15]. Comparisons between Colias and Melitaea cinixia butterfly Pgi nuclear polymorphisms indicate convergent evolution [18]. Increased Colias survival [14] and M. cinixia population growth rates [16] have both been correlated with Pgi allelic variants. This variation is likely a product of long-term balancing selection on charge-changing amino acid substitutions on external loops of the protein’s surface [17, 18]. The consistent association of Pgi polymorphisms with variation in individual performance, coupled with no known duplication events in arthropods, has led to this locus being proposed as a general adaptive marker for conservation genetics in this phylum, akin to MHC in vertebrates [13].
Flight [22] and adaptation to low temperature [23] are both linked with varying energy demands and increased oxidative stress. The metabolic rate of insects during flight may increase by over a 100-fold compared with when they are at rest [22]. This metabolically-demanding activity results in increased oxidative stress, which causes cellular damage and even increased mortality in houseflies (Musca domestica) [24]. Furthermore, the increased energy demand of flight may require the insect to exploit different fuels, as seen in the flight-capable morphs of the wing-polymorphic cricket (Gryllus firmus), where winged individuals increase lipid biosynthesis in preparation for flight [25]. Similar patterns are observed in insects exposed to low temperatures, where depressed metabolic rates result in the accumulation of reactive oxygen species [23] and there is increased lipid biosynthesis in preparation for overwintering in several insect species [26]. One way to test the impact of varying metabolic demands on the evolution of glycolysis is to study variation in these component genes across related taxa that have evolved to new, potentially stressful lifestyles, and that vary in their ability to fly.
The Australasian Lanceocercata stick insects (Order: Phasmatodea) form a monophyletic clade [27–30] comprising species from five subfamilies, Pachymorphinae, Tropidoderinae, Xeroderinae, Phasmatinae, Eurycanthinae, and Platycraninae, with distinct morphological characters [28]. The Lancerocercata have undergone an impressive evolutionary radiation over the last 50 Mya [29]. This includes the convergent evolution of different body forms [30], loss of flight in one or both sexes, shifts in diet, and adaptation to low-temperature environments in New Zealand, including four wingless species found in high alpine areas where they regularly experience sub-zero temperatures [31, 32]. These contrasting lifestyles within Lanceocercata are expected to afford a diverse range of metabolic demands. This diversity provides a comprehensive model system in which to determine whether glycolysis has been the target of positive selection, and if so, at which steps and even at which sites within these enzymes selection has occurred. To pinpoint positive selection within Lanceocercata we obtained partial cDNA sequences of seven core glycolytic enzymes, together with the enzymes immediately before and after the pathway, across a phylogeny of 29 Lanceocercata species. We found that the Pgi gene has been duplicated within Lanceocercata, with at least two of the three paralogous copies and Gapdh evolving under positive selection.
Results
DNA sequencing of genes from glycolysis
We used de novo transcriptomes derived from 454 sequencing of three New Zealand species (Micrarchus nov. sp. 2 [31], Niveaphamsa annulata and Clitarchus hookeri) to design PCR primers and amplify 19 genes to reconstruct the evolutionary history of 29 Lanceocercata stick insects. Of these partial gene transcripts, ten were distributed across the glycolytic pathway, including seven of the ten core enzymes (Figure 1). In addition, six nuclear protein coding genes with a variety of cellular functions (Calr, Ef1a, Flnc, Gsk3b, Mer and Sord), two mitochondrial genes (COI, COII), and 28S rDNA were sequenced from cDNA for phylogenetic reconstruction.
Positive selection in the glycolytic pathway. Galm and Pdh are not part of the core glycolysis genes but operate in the steps immediately before or after the pathway. Genes in Bold were sequenced in this study, with ω denoting positive selection having been inferred at individual codons. Branch points to and from the pentose phosphate pathway are included.
Pgi duplications
Although only a single copy of Pgi has been previously characterised in arthropods, the transcriptome assemblies revealed three paralogous copies of Pgi within Lanceocercata, with the duplication events presumably occurring after the split between Timema and all other Phasmatodea (Additional file 1: Figure S1), which occurred more than 95 Myr ago [29]. Sequences of the full length open reading frame from the transcriptomes show extensive predicted amino acid variation within and among Pgi paralogues (Additional file 1: Figure S2), with the percentage of identical amino acid sites within the three species being higher for pairwise comparisons among orthologues (mean = 94.2%) than paralogues (mean = 70.2%). Subsequent PCR amplification of Pgi paralogues across a wider range of species was possible from a region within Pgi1 (675 bp) and Pgi2 (1452 bp), but not from Pgi3 due to the lack of unique primer binding sites.
Lanceocercata phylogeny reconstruction
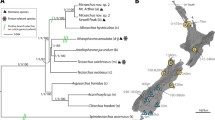
To infer evolutionary relationships among the sampled Lanceocercata species, a combined Bayesian phylogeny (Figure 2) was constructed from a 15,034 bp partitioned alignment of the 18 protein coding genes and 28S. An average of 13,990 bp of sequence data was used per species, with the minimum of 9,692 bp (Gen. Nov. 1, sp. nov. 1) and a maximum of 15,034 bp (Acanthoxyla sp. Argosarchus horridus, Asteliaphasma jucundum, all Clitarchus sp., Pseudoclitarchus sentus, Tectarchus ovobessus and T. salebrosus). This combined phylogeny has variable support for a monophyletic clade containing all New Zealand species. The different lifestyles of flight and alpine habitation are scattered across the phylogeny.
Lanceocercata phylogeny with codons under selection. Combined Bayesian Lanceocercata phylogenetic tree constructed using 15,077 bp from 19 genes with branch lengths proportional to the estimated number of substitutions per site. Nodal support values are nucleotide posterior probabilities/ nucleotide maximum likelihood bootstraps/ amino acid posterior probabilities and /amino acid maximum likelihood bootstraps. Asterisks denote nodes with 100% support; Dashes indicate nodes with less than 50% support. Amino acids sites identified by the M8 Bayes Empirical Bayes (BEB) analysis are depicted on the right with polarity and hydrophobicity highlighted.
Codons under positive selection in the glycolytic pathway
The selective pressures acting on a protein-coding DNA sequence can be inferred from the ratio of nonsynonymous to synonymous nucleotide substitutions (ω). Purifying selection is indicated by ω <1, whereas ω =1 implies neutral evolution and ω >1 indicates positive selection [33]. Typically, selection acts on relatively few amino acid sites within a protein-coding gene. As a result, standard neutrality tests that average ω across the entire reading frame often fail to infer incidences of positive selection [34]. Amino acid residues likely to have been influenced by positive selection within a protein can be identified by estimating ω with site-based maximum likelihood methods [35], even though this approach may be overly conservative [36]. Positive selection can be inferred by comparing the goodness of fit for a set of nested models. Models that allow ω to vary above one to include the effect of positive selection are compared with null models where ω is not allowed to fluctuate above 1, meaning the observed nucleotide variation is solely a product of purifying selection and neutrality. Likelihood ratio tests and chi-square distributions are used to assess the statistical significance between the models.
Site-based likelihood models were used to infer positive selection acting on ten Lanceocercata genes associated with the glycolytic pathway (Figure 1). We found that all glycolysis genes are predominantly evolving under the influence of strong purifying selection when a single value of ω is calculated across the protein coding sequences using the one-ratio (M0) model (mean ω = 0.10; range 0.04-0.19; Table 1). However, there is significant among-site variation in ω within all genes, with the M3 model permitting three ω values, providing a significantly better fit to the data than the M0 model (p value <0.001; d.f. = 4; M0:M3; Table 1; Additional file 1: Table S1). To test for positive selection the M7 beta neutral model with eight categories of ω, all less than one and together comprising a beta distribution, is compared with the M8 model that has one additional ω value that is free to vary above one and therefore account for positive selection. The M8 model is a significantly better fit for three core glycolysis genes (Pgi1 (p < 0.001), Pgi2 (p < 0.001), Gapdh (p < 0.05) and aldose 1-epimerase (Galm (p < 0.05); d.f. = 2), which links glycolysis to the Leloir pathway, indicating positive selection acts on these genes (Table 1; Additional file 1: Table S1). A more conservative likelihood ratio test for positive selection replaces the M7 null model with M8a, which has an extra category of ω fixed at one. The M8:M8a nested model comparison identifies positive selection in Pgi1 (p < 0.001) and Pgi2 (p < 0.05) (d.f. = 50:50 mixture of point mass 0 and 1; Additional file 1: Table S1). The most stringent approach to identify positive selection compares the M1a null model, which has two values of ω (ω <1, ω =1), and M2a, which has an additional third category of ω that is greater than one. This approach still identifies positive selection in Pgi1 (p < 0.001; d.f. = 2; Additional file 1: Table S1), suggesting that this inference is especially robust.
Amino acid sites under positive selection in the glycolytic pathway
Four genes had sites identified under positive selection in the nested M7:M8 site models. A subsequent Bayes Empirical Bayes (BEB) method implemented in PAML was used to assess the significance of the ω ratio at each of these positively selected codon positions. In total six codons were identified as under significant positive selection (Posterior probability (P) > 95%; Table 2). These include one site in Gapdh (Asn63), two in Pgi1 (Val414 and Lys454) and three in Pgi2 (Asn131, Asn324 and Gly425). All these codons involve predicted amino acid substitutions that vary in residue polarity and hydrophobicity, apart from Asn63 in Gapdh, where only hydrophobicity is altered (Figure 2). The two Pgi paralogues share a similar pattern of BEB posterior probabilities (Figure 3), with regions of high posterior probability (P > 0.75) being predominately located in disordered regions of the protein secondary structure (Pgi1 = 3 out of 4; Pgi2 = 7 out of 11; Figure 3) as opposed to well-structured α-helices (Pgi1 = 1 out of 4; Pgi2 = 4 out of 11) or β-strands (Pgi1 = 0 out of 4; Pgi2 = 0 out of 11). Disordered regions are defined as areas that in isolation do not fold into well-defined 3D structures and conversely sample a large portion of the available conformational space, including features such as loops and extended domains [37]. In other arthropod species, sites identified as either under positive selection, balancing selection, associated with fitness differences or segregating between species in Pgi also predominately occur within these disordered regions (Colias sp.: 18 out of 27 [17]; Melitaea cinixia: 4 out of 5 [18]; Tigriopus californicus: 4 out of 6 [38]. Similar patterns of high posterior probabilities of positive selection are seen among paralogues and orthologues (Figure 3). There is direct overlap between Val414 in Pgi1, which is under positive selection (P = 0.999), and the homologous site in Pgi2 (P = 0.836). Additionally, Asn131, which is under positive selection in Pgi2 (P = 0.98) is homologous to a segregating site between Colias sp. (Gly128) [17].
Positive selection within Pgi and Gapdh . Distribution of positively selected sites across three Lanceocercata stick insect genes: phosphoglucose isomerase 1 (Pgi1), phosphoglucose isomerase 2 (Pgi2) and glyceraldehyde 3-phosphate dehydrogenase (Gapdh); Probability of each codon position being under positive selection (ω > 1) using Bayes Empirical Bayes analysis under the M8 site-specific model in PAML. The Pgi1 analysis consists of 225 codons from 28 species, Pgi2 484 codons from 20 species and Gapdh 279 codons from 29 species. Codon position based on full-length alignment of both Pgi genes from Micrarchus nov. sp. 2. Amino acid sites identified in previous studies as either under positive selection, associated with fitness differences or segregating between species for PGI are shown. The secondary structure of each protein in relation to codon position is shown.
High resolution crystal structures of the glycolytic enzymes are available, allowing the location of codons identified as under positive selection by the BEB analysis to be visualised in a structural context. Predicted structural models of full length Micrarchus nov. sp. 2 protein sequences were generated using SWISS-MODEL [39]. PGI1 and PGI2 were modelled against the pig PGI structure [40] (1GZD; E = 0; sequence identity = 70 & 66% respectively), whereas Gapdh was modelled against the spiny blue lobster GAPDH structure [41] (1DSS; E = 1.71e-146; sequence identity = 81%). Five of six residues under positive selection in these enzymes are located on the surface of the protein (Figure 4). None of the sites under positive selection are closely associated with the conserved active site residues (Figure 4).
Discussion
Here we infer positive selection within the glycolytic pathway of Australasian Lanceocerata stick insects encompassing a wide diversity of lifestyles and habitats. The genes under positive selection are not key metabolic control steps regulating flux through the glycolytic pathway. Across the pathway, Pgi and Gapdh exhibit the strongest signal for positive selection using the conservative codon-based likelihood methods. This may be attributed to their positions in the glycolysis pathway and association with branch points diverting flux through the pentose phosphate pathway (PPP) (Figure 1).
The PPP synthesises NADPH for lipid biosynthesis and to neutralise reactive oxygen species accumulated during periods of oxidative stress [1]. Pgi allelic variants and GAPDH inactivation are respectively known to differentially regulate flux through the PPP in the sea anemone Metridium senile[2] and C. elegans[1]. Excess NADPH synthesised by the PPP may play an important role in adaptation to novel stressful life histories by allowing insects either to exploit different energy storage pools, or to compensate for increased oxidative stress, or both. Increased lipid biosynthesis is documented in overwintering insects [26] and winged morphs of Gryllus firmus[25]. Insects are also particularly susceptible to oxidative stress due to: (i) their tracheal respiratory system; (ii) herbivorous diets being rich in pro-oxidants [42]; (iii) extreme demand for oxygen during flight [22]; and (iv) exposure to low-temperature [23].
The amino acid substitutions at the sites inferred to be under positive selection do not discretely partition among different life history strategies. The lack of a clear pattern may in part be due to the complexity of their life-history traits. For example, alpine species may experience selection for vastly different metabolic regimes depending on whether they are freeze tolerant or freeze avoidant [43]. Understanding how adaptation to new stressful lifestyles alters flux through both the glycolytic and PPP will require integrated biochemical enzyme assays in relation to sites under positive selection [44, 45], and will ultimately indicate the relative importance of maintaining energy production against exploiting the different energy storage pools and reducing the cost of stress induced oxidative damage in adaptation.
The continued association of Pgi with an array of fitness measures has led to the suggestion that this locus could be used as an adaptive marker in arthropods [13]. However, the occurrence of paralogues affects its practical implementation using traditional PCR-based methods, at least in the Euphasmatodea. Gene duplications may become fixed if there is existing balancing selection, as the fixation of both original alleles removes the possibility of the deleterious occurrence of homozygotes at the original locus [46]. The Pgi duplications in Lanceocercata are possibly a consequence of existing balancing selection preceding subsequent gene duplication. Gene duplications may eventually lead to subfunctionalisation of tissue-specific forms of the protein, as seen in teleost fish Pgi[47] and Tpi[48].
The positive selection acting on Pgi1 and Pgi2 results in changes to the polarity and hydrophobicity of the translated amino acids. The charge-changing amino acid substitutions in the stick insect PGI proteins may cause biochemical and fitness differences similar to those observed in Colias PGI [17, 18]. In both stick insects and Colias, a majority of these sites occur in disordered regions of the PGI secondary structure, in regions that are more conformationally-flexible than the α-helices and β-strands. These intrinsically-disordered protein regions are thought to tolerate increased genetic variation and are important for maintaining adaptive potential [37, 49]. Furthermore, protein function is maintained by purifying selection against internal changes that may disrupt enzyme function, restricting positive selection to amino acids exposed to the surface of the protein’s three dimensional structure [50]. Relatively few modifications are required for environment-specific adaptations to develop, as seen in vertebrate rhodopsin where 94% of amino acid sites are evolving neutrally [36], and in Mytilus mussels where temperature adaptation in isocitrate dehydrogenase is driven by two amino acid substitutions [51]. While surface modifications can alter catalytic performance of enzymes, as seen in lactate dehydrogenase [52], they are more likely to affect a protein’s thermal stability [53]. A similar pattern of positive selection across the overlapping protein coding sequence is observed in the Pgi paralogues investigated here. There is also correspondence in the position of sites under positive directional selection in the stick insects and those identified as either under positive selection, associated with fitness differences or segregating between other arthropod species ([18, 38, 54]; Figure 3). A majority of the Pgi coding sequence is conserved by strong purifying selection maintaining protein function. However, positive selection appears to be able to act on specific portions of the Pgi gene that are: (i) intrinsically disordered regions of the protein structure where genetic variation is maintained; and (ii) surface residues that can influence enzyme conformation and efficiency. Modifications within these convergent windows of positive selection are presumably responsible for driving enzyme properties towards local optima in response to a host of species-specific adaptations.
Conclusion
These results from Lanceocercata stick insects illustrate that adaptations within metabolic pathways may be a delicate trade-off between maintaining energy production, optimally exploiting different energy pools, and reducing the effects of increased oxidative stress in the system. Maintaining this balance affects organisms regardless of the adaptation, and selection likely targets enzymes at branch points due to their pleiotropic effects on flux through metabolic pathways. The complexity of enzyme and pathway interactions requires further integrative studies to understand how the metabolic network evolves as a whole.
Methods
Lanceocercata specimens were collected from Australia, New Caledonia and New Zealand (Additional file 1: Table S3). Samples were preserved in ethanol and stored at –20°C or directly snap frozen and stored at –80°C before RNA extraction. Total RNA was extracted from leg muscle using the RNeasy Mini kit (Qiagen) from 55 individuals representing 29 species. First strand cDNA synthesis was random hexamer primed using the SuperScript III First Strand Kit (Invitrogen) following the manufacturer’s protocol. cDNA libraries were subsequently amplified used 5 μL first strand cDNA, 5 μL of 2 mM dNTP (Roche), 10 μL of 10X FastStart Taq DNA Polymerase PCR Buffer with MgCl2 (Roche),1 μL of 15 mM MgCl2 (Roche), 2 U FastStart Taq DNA Polymerase (Roche) and 5 μL of 25 μM random hexamer in a total volume of 53 μL. Amplifications were performed on a GeneAmp PCR system 9700 thermal cycler (Applied Biosystems) using the following parameters: 5 min at 95°C, 3 min at 50°C , 40 sec 72°C; 40 cycles of 40 sec at 94°C,40 sec at 65°C and 40 sec at 72°C; and 10 min at 72°C.
Sequencing of glycolysis genes
Primers to amplify partial gene sequences for 19 genes were designed from three 454 de novo transcriptomes of three New Zealand species (Micrarchus nov. sp. 2 [31], Niveaphamsa annulata and Clitarchus hookeri) using Primer3 [55] in Geneious v.5.6 [56]. The N. annulata and C. hookeri transcriptomes were constructed from previously published methods [31] and consisted of 276 K reads from eight individuals and 235 K reads from six individuals, respectively (NCBI SRA submission SRA061479 & SRA061480). Primers were designed to amplify parts of ten gene transcripts (Eno, Galm, Gapdh, Pdh, Pgi1, Pgi2, Pgk, Pglym, Pyk, and Tpi) distributed across the glycolytic pathway. In addition, six nuclear protein coding genes with a variety of cellular functions (Calr, Ef1a, Flnc, Gsk3b, Mer and Sord), two mitochondrial genes (COI, COII), and 28S rDNA were sequenced to assist with phylogeny reconstruction. Target genes were PCR amplified with reactions consisting of approximately 5 ng cDNA, 2.5 μL of 2 mM dNTP (Roche), 1 μL of 10 μM BSA (Sigma-Aldrich), 2.5 μL of 10X FastStart Taq DNA Polymerase PCR Buffer with MgCl2 (Roche), 1.5 U FastStart Taq DNA Polymerase (Roche) and 7.25 pmol of the forward and reverse primers (Sigma-Aldrich) in a total volume of 25 μL. Amplifications were performed on a GeneAmp PCR system 9700 thermal cycler (Applied Biosystems) using the following parameters: 5 min at 95°C; 35 cycles of 1 min at 94°C,1 min at primer specific annealing temperature and 1 min at 72°C; and 10 min at 72°C (Primer sequences and annealing temperature in Additional file 1: Table S2). Successful amplification was assessed by gel electrophoresis on 1% agarose gels containing 0.5 mg mL–1 ethidium bromide.
PCR products were sequenced using BigDye Terminator Cycle Sequencing Ready Reaction Mix v3.1 (Applied Biosystems). Cycle sequencing products were cleaned using the BigDye Xterminator Purification Kit (Applied Biosystems) and analysed in both directions on the ABI Prism 3100 Genetic Analyzer (Applied Biosystems). Sequences were subsequently cleaned, trimmed and aligned using Geneious v.5.6. To verify nucleotide substitutions were not a result of polymerase errors, the generated sequences were compared with the High-Throughput Sequencing generated data; and intraspecifically if multiple samples were available for a species. Three core glycolytic enzymes were not characterised, due to multiple expressed copies (hexokinase) or insufficient sequence information in the original transcriptomes (phosphofructokinase and aldose). Sequences were submitted to the NCBI Genbank repository (accession KF383428 to KF383954).
Subsequent sequence analyses only used one sequence per species selected on the tiered criteria of: sequence length; fewest variable nonsynonymous sites; fewest synonymous sites; greatest agreement to consensus; and finally, using the individual selected for a majority of other genes. If necessary, haplotype reconstruction of polymorphic sequences was performed using SeqPHASE [57] and PHASE v2.1 [58, 59] (probability of > 90%) prior to calculating descriptive statistic in DnaSP v.5 [60]. Substitution models for nucleotide and amino acid alignments were selected using the corrected Akaike information criterion [61] from jModelTest v.0.1.1 [62, 63] and ProtTest3 v.3.2 [63, 64] respectively. A Bayesian phylogeny for the 15,034 bp partitioned alignment using MrBayes v.3.2 [65, 66] run for 10,000,000 generations. Additional phylogeny nodal support values were obtained by maximum likelihood (100 bootstraps) in Garli v.2.0 [67].
4.5.2 Inferring selection
To screen for positive selection in the ten glycolysis protein coding genes, ω was estimated by maximum likelihood using site models implemented in the CODEML program of PAML v.4.5 [33]. The models implemented (M0, M1a, M2a, M3, M7, M8 and M8a) are extensively described elsewhere [33]. Complex models (M1a, M2a, M3, M7, M8, M8a) permit more than one category of ω, allowing individual codons to be identified as under positive selection (>1) when the average ω across the whole gene indicates purifying selection (<1). Likelihood ratio tests (LRTs) between models where ω is allowed to vary above one, and the associated null models where ω is fixed at one, allow inference on the selective pressure acting on the sequence [33]. Codons under positive selection were identified using BEB method under the M8 model. Secondary protein structures were predicted using PSIPRED V.3.3 [68]. Three-dimensional structural modelling of genes identified as under positive selection was performed using SWISS-MODEL [39] and visualised in PyMOL v.0.99 [69].
Availability of supporting data
The data sets supporting the results of this article are available in the National Center for Biotechnology Information GenBank (KF383428 to KF383954) and Sequence Read Archive (SRA061479 and SRA061480) repositories.
References
Ralser M, Wamelink M, Kowald A, Gerisch B, Heeren G, Struys E, Klipp E, Jakobs C, Breitenbach M, Lehrach H, Krobitsch S: Dynamic rerouting of the carbohydrate flux is key to counteracting oxidative stress. J Biol. 2007, 6: 10-10.1186/jbiol61.
Zamer WE, Hoffmann RJ: Allozymes of glucose-6-phosphate isomerase differentially modulate pentose-shunt metabolism in the sea anemone Metridium senile. Proc Natl Acad Sci U S A. 1989, 86: 2737-2741. 10.1073/pnas.86.8.2737.
Eanes WF: Molecular population genetics and selection in the glycolytic pathway. J Exp Biol. 2011, 214: 165-171. 10.1242/jeb.046458.
Kacser H, Burns JA: The molecular basis of dominance. Genetics. 1981, 97: 639-666.
Hartl DL, Dykhuizen DE, Dean AM: Limits of adaptation: the evolution of selective neutrality. Genetics. 1985, 111: 655-674.
Eanes WF: Analysis of selection on enzyme polymorphisms. Annu Rev Ecol Syst. 1999, 30: 301-326. 10.1146/annurev.ecolsys.30.1.301.
Watt WB, Dean AM: Molecular-functional studies of adaptive genetic variation in prokaryotes and eukaryotes. Annu Rev Genet. 2000, 34: 593-622. 10.1146/annurev.genet.34.1.593.
Wright KM, Rausher MD: The evolution of control and distribution of adaptive mutations in a metabolic pathway. Genetics. 2010, 184: 483-502. 10.1534/genetics.109.110411.
Rausher MD: The evolution of genes in branched metabolic pathways. Evolution. 2013, 67: 34-48. 10.1111/j.1558-5646.2012.01771.x.
Eanes WF, Merritt TJS, Flowers JM, Kumagai S, Sezgin E, Zhu C-T: Flux control and excess capacity in the enzymes of glycolysis and their relationship to flight metabolism in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2006, 103: 19413-19418. 10.1073/pnas.0607095104.
Flowers JM, Sezgin E, Kumagai S, Duvernell DD, Matzkin LM, Schmidt PS, Eanes WF: Adaptive evolution of metabolic pathways in Drosophila. Mol Biol Evol. 2007, 24: 1347-1354. 10.1093/molbev/msm057.
Riddoch BJ: The adaptive significance of electrophoretic mobility in phosphoglucose isomerase (PGI). Biol J Linn Soc. 1993, 50: 1-17. 10.1111/j.1095-8312.1993.tb00915.x.
Wheat CW: Phosphoglucose isomerase (Pgi) performance and fitness effects among Arthropods and its potential role as an adaptive marker in conservation genetics. Conserv Genet. 2010, 11: 387-397. 10.1007/s10592-009-0042-7.
Watt WB: Adaptation at specific loci. I. Natural selection on phosphoglucose isomerase of Colias butterflies: biochemical and populations aspects. Genetics. 1977, 87: 177-194.
Dahlhoff EP, Rank NE: Functional and physiological consequences of genetic variation at phosphoglucose isomerase: heat shock protein expression is related to enzyme genotype in a montane beetle. Proc Natl Acad Sci U S A. 2000, 97: 10056-10061. 10.1073/pnas.160277697.
Hanski I, Saccheri I: Molecular-level variation affects population growth in a butterfly metapopulation. PLoS Biol. 2006, 4: e129-10.1371/journal.pbio.0040129.
Wheat CW, Watt WB, Pollock DD, Schulte PM: From DNA to fitness differences: sequences and structures of adaptive variants of Colias phosphoglucose isomerase (PGI). Mol Biol Evol. 2006, 23: 499-512.
Wheat CW, Haag CR, Marden JH, Hanski I, Frilander MJ: Nucleotide polymorphism at a gene (Pgi) under balancing selection in a butterfly metapopulation. Mol Biol Evol. 2010, 27: 267-281. 10.1093/molbev/msp227.
Dykhuizen DE, Hartl DL: Functional effects of PGI allozymes in Escherichia coli. Genetics. 1983, 105: 1-18.
Hall JG: The adaptation of enzymes to temperature: catalytic characterization of glucosephosphate isomerase homologues isolated from Mytilus edulis and Isognomon alatus, bivalve molluscs inhabiting different thermal environments. Mol Biol Evol. 1985, 2: 251-269.
Filatov DA, Charlesworth D: DNA polymorphism, haplotype structure and balancing selection in the Leavenworthia PgiC Locus. Genetics. 1999, 153: 1423-1434.
Krogh A, Weis-Fogh T: The respiratory exchange of the desert locust (Schistocerca gregaria) before, during and after flight. J Exp Biol. 1951, 28: 344-357.
Lalouette L, Williams CM, Hervant F, Sinclair BJ, Renault D: Metabolic rate and oxidative stress in insects exposed to low temperature thermal fluctuations. Comp Biochem Physiol A Mol Integr Physiol. 2011, 158: 229-234. 10.1016/j.cbpa.2010.11.007.
Yan L, Sohal R: Prevention of flight activity prolongs the life span of the housefly, Musca domestica, and attenuates the age-associated oxidative damamge to specific mitochondrial proteins. Free Radic Biol Med. 2000, 29: 1143-1150. 10.1016/S0891-5849(00)00423-8.
Zera A, Larsen A: The metabolic basis of life history variation: genetic and phenotypic differences in lipid reserves among life history morphs of the wing-polymorphic cricket, Gryllus firmus. J Insect Physiol. 2001, 47: 1147-1160. 10.1016/S0022-1910(01)00096-8.
Downer R, Matthews J: Patterns of lipid distribution and utilisation in insects. Am Zool. 1976, 16: 733-745.
Whiting MF, Bradler S, Maxwell T: Loss and recovery of wings in stick insects. Nature. 2003, 421: 264-267. 10.1038/nature01313.
Bradler S: Phylogeny of the stick and leaf insects (Insecta: Phasmatodea). Species, Phylogeny and Evolution. 2009, 2: 3-139.
Buckley TR, Nylander JAA, Bradler S: The phylogenetic placement and biogeographical origins of the New Zealand stick insects (Phasmatodea). Syst Entomol. 2010, 35: 207-225. 10.1111/j.1365-3113.2009.00505.x.
Buckley TR, Attanayake D, Bradler S: Extreme convergence in stick insect evolution: phylogenetic placement of the Lord Howe Island tree lobster. Proc R Soc B Biol Sci. 2009, 276: 1055-1062. 10.1098/rspb.2008.1552.
Dunning LT, Dennis AB, Park D, Sinclair BJ, Newcomb RD, Buckley TR: Identification of cold-responsive genes in a New Zealand alpine stick insect using RNA-Seq. Comparative Biochemistry and Physiology Part D: Genomics and Proteomics. 2013, 8: 24-31. 10.1016/j.cbd.2012.10.005.
Dennis AB, Dunning LT, Dennis C, Sinclair BJ, Buckley TR: Overwintering in New Zealand stick insects. New Zealand Entomologist. 2013, http://www.tandfonline.com/doi/abs/10.1080/00779962.2013.784235,
Yang Z: PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007, 24: 1586-1591. 10.1093/molbev/msm088.
Yang Z, Nielsen R: Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol. 2002, 19: 808-817.
Lassance J-M, Liénard MA, Antony B, Qian S, Fujii T, Tabata J, Ishikawa Y, Löfstedt C: Functional consequences of sequence variation in the pheromone biosynthetic gene pgFAR for Ostrinia moths. Proc Natl Acad Sci U S A. 2013, 110: 3967-3972. 10.1073/pnas.1208706110.
Yokoyama S, Tada T, Zhang H, Britt L: Elucidation of phenotypic adaptations: molecular analyses of dim-light vision proteins in vertebrates. Proc Natl Acad Sci U S A. 2008, 105: 13480-13485. 10.1073/pnas.0802426105.
Schlessinger A, Schaefer C, Vicedo E, Schmidberger M, Punta M, Rost B: Protein disorder - a breakthrough invention of evolution?. Curr Opin Struct Biol. 2011, 21: 412-418. 10.1016/j.sbi.2011.03.014.
Schoville SD, Flowers JM, Burton RS: Diversifying selection underlies the origin of allozyme polymorphism at the phosphoglucose isomerase locus in Tigriopus californicus. PLoS ONE. 2012, 7: e40035-10.1371/journal.pone.0040035.
Schwede T, Kopp J, Guex N, Peitsch MC: SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003, 31: 3381-3385. 10.1093/nar/gkg520.
Davies C, Muirhead H: Crystal structure of phosphoglucose isomerase from pig muscle and its complex with 5-phosphoarabinonate. Proteins: Structure, Function, and Bioinformatics. 2002, 49: 577-579. 10.1002/prot.10255.
Song S, Xu Y, Lin Z, Tsou C: Structure of active site carboxymethylated d-glyceraldehyde-3-phosphate dehydrogenase from Palinurus versicolor. J Mol Biol. 1999, 287: 719-725. 10.1006/jmbi.1999.2628.
Ahmad S: Biochemical defence of pro-oxidant plant allelochemicals by herbivorous insects. Biochem Syst Ecol. 1992, 20: 269-296. 10.1016/0305-1978(92)90040-K.
Voituron Y, Mouquet N, de Mazancourt C, Clobert J: To freeze or not to freeze? An evolutionary perspective on the cold-hardiness strategies of overwintering ectotherms. Am Nat. 2002, 160: 255-270. 10.1086/341021.
Storz JF, Wheat CW: Integrating evolutionary and functional approaches to infer adaptation at specific loci. Evolution. 2010, 64: 2489-2509. 10.1111/j.1558-5646.2010.01044.x.
Zera AJ: Microevolution of intermediary metabolism: evolutionary genetics meets metabolic biochemistry. J Exp Biol. 2011, 214: 179-190. 10.1242/jeb.046912.
Proulx SR, Phillips PC: Allelic divergence precedes and promotes gene duplication. Evolution. 2006, 60: 881-892.
Sato Y, Nishida M: Post-duplication charge evolution of phosphoglucose isomerases in teleost fishes through weak selection on many amino acid sites. BMC Evol Biol. 2007, 7: 204-10.1186/1471-2148-7-204.
Merritt TJS, Quattro JM: Evidence for a period of directional selection following gene duplication in a neurally expressed locus of triosephosphate isomerase. Genetics. 2001, 159: 689-697.
Nilsson J, Grahn M, Wright A: Proteome-wide evidence for enhanced positive Darwinian selection within intrinsically disordered regions in proteins. Genome Biol. 2011, 12: R65-10.1186/gb-2011-12-7-r65.
Bustamante CD, Townsend JP, Hartl DL: Solvent accessibility and purifying selection within proteins of Escherichia coli and Salmonella enterica. Mol Biol Evol. 2000, 17: 301-308. 10.1093/oxfordjournals.molbev.a026310.
Lockwood BL, Somero GN: Functional determinants of temperature adaptation in enzymes of cold- versus warm-adapted mussels (Genus Mytilus). Mol Biol Evol. 2012, 29: 3061-3070. 10.1093/molbev/mss111.
Fields PA, Somero GN: Hot spots in cold adaptation: localized increases in conformational flexibility in lactate dehydrogenase A4 orthologs of Antarctic notothenioid fishes. Proc Natl Acad Sci U S A. 1998, 95: 11476-11481. 10.1073/pnas.95.19.11476.
Fields PA: Protein function at thermal extremes: balancing stability and flexibility. Comp Biochem Physiol A Mol Integr Physiol. 2001, 129: 417-431. 10.1016/S1095-6433(00)00359-7.
Watt WB, Wheat CW, Meyer EH, Martin JF: Adaptation at specific loci. VII. Natural selection, dispersal and the diversity of molecular-functional variation patterns among butterfly species complexes (Colias: Lepidoptera, Pieridae). Mol Ecol. 2003, 12: 1265-1275. 10.1046/j.1365-294X.2003.01804.x.
Rozen S, Helen J: Primer3 on the WWW for general users and for biologist programmers. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Edited by: Krawetz S, Misener S. 2000, Totowa, NJ: Humana Press, 365-386.
Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, Field M, Heled J, Kearse M, Markowitz S, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A: 2012, Geneious v5.6, available form URL http://www.geneious.com/
Flot JF: Seqphase: a web tool for interconverting phase input/output files and fasta sequence alignments. Mol Ecol Resour. 2010, 10: 162-166. 10.1111/j.1755-0998.2009.02732.x.
Stephens M, Scheet P: Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet. 2005, 76: 449-462. 10.1086/428594.
Stephens M, Donnelly P: A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003, 73: 1162-1169. 10.1086/379378.
Librado P, Rozas J: DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009, 25: 1451-1452. 10.1093/bioinformatics/btp187.
Hurvich CM, Tsai C-L: Regression and time series model selection in small samples. Biometrika. 1989, 76: 297-307. 10.1093/biomet/76.2.297.
Posada D: jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008, 25: 1253-1256. 10.1093/molbev/msn083.
Guindon S, Gascuel O: A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003, 52: 696-704. 10.1080/10635150390235520.
Darriba D, Taboada GL, Doallo R, Posada D: ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011, 27: 1164-1165. 10.1093/bioinformatics/btr088.
Ronquist F, Huelsenbeck JP: MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003, 19: 1572-1574. 10.1093/bioinformatics/btg180.
Huelsenbeck JP, Ronquist F: MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001, 17: 754-755. 10.1093/bioinformatics/17.8.754.
Zwickl DJ: Unpublished Ph.D. dissertation. Genetic Algorithm Approaches for the Phylogenetic Analysis of Large Biological Sequence Datasets Under the Maximum Likelihood Criterion. 2006, Austin, TX: The University of Texas
Buchan DWA, Ward SM, Lobley AE, Nugent TCO, Bryson K, Jones DT: Protein annotation and modelling servers at University College London. Nucleic Acids Res. 2010, 38: W563-W568. 10.1093/nar/gkq427.
Delano WL: The PyMOL Molecular Graphics System. 2002, San Carlos, California: DeLano Scientific
Acknowledgements
We thank Rich Leschen, Shelly Myers, Litza Coello, Mhairi McFarlane, Ana Ramon-Laca, Lucy Shield, and Clive Williams for assistance with fieldwork, Thomas Merritt for insightful comments on the manuscript, the Department of Conservation for access to field sites, and members of the ecological genetics lab, Landcare Research. We would also like to thank the editor and the two anonymous reviewers for their insightful comments. The authors wish to acknowledge the contribution of the New Zealand eScience Infrastructure (NeSI) high-performance computing facilities and the staff at the Centre for eResearch at the University of Auckland. Funding was provided by The Royal Society of New Zealand Marsden Fund (LCR0901), the Allan Wilson Centre for Molecular Ecology and Evolution and Core funding for Crown Research Institutes from the New Zealand Ministry of Business, Innovation and Employment’s Science and Innovation Group.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contribution
LTD and GT sequenced target genes; AD provided additional transcriptome data; LTD Preformed Selection analyses; LTD, AD, BJS, RDN and TRB conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
12862_2013_2447_MOESM1_ESM.docx
Additional file 1: Table S1: Likelihood parameter values for site models. Table S2. Primer sequences and annealing temperatures. Table S3. Species samples collection details. Figure S1. Bayesian arthropod PGI phylogeny. Figure S2. Amino acid alignment of all three PGI paralogues. (DOCX 911 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Dunning, L.T., Dennis, A.B., Thomson, G. et al. Positive selection in glycolysis among Australasian stick insects. BMC Evol Biol 13, 215 (2013). https://doi.org/10.1186/1471-2148-13-215
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2148-13-215