Abstract
Polycystic ovary syndrome (PCOS) is a life-long reproductive, endocrine, and metabolic disorder that affects up to 17% of women of reproductive age. However, the effect of granulosa cells (GCs) transcriptome changes on oocyte capacity and follicular development in patients with PCOS has not been elucidated. This study aims to analyze transcriptome changes in GCs of PCOS from different perspectives and explore potential biomarkers for the diagnosis and treatment of PCOS. The gene expression profiles of GSE34526 and GSE107746 were obtained from the GEO database. Differentially expressed genes (DEGs) and key signaling pathways were identified. Gene Set Enrichment Analysis (GSEA) revealed that Toll-like receptors, NOD-like receptors, and NOTCH signaling pathways were obviously enriched in GCs of PCOS. We further analyzed DEGs from three aspects: transcription factors (TFs), secreted proteins, and follicular development. Compared with normal GCs, the DEGs encoding TFs and secretory proteins in GCs of PCOS remarkably changed. Besides, HAS2 and CBLN1, which are highly expressed in preovulatory follicular GCs and may trigger ovulation, were significantly decreased in GCs of PCOS. This study found candidate genes and signaling pathways in PCOS, providing new insights and foundations for the etiology of PCOS. Besides, HSA2 and CBLN1 may be potential therapeutic biomarkers for ovulation disorders in PCOS.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Polycystic ovary syndrome (PCOS) is a life-long reproductive, endocrine, and metabolic disorder that affects up to 17% of women of reproductive age and is characterized as ovulatory dysfunction, hyperandrogenemia, and polycystic ovaries [1]. With oligoovulation or anovulation, PCOS is the leading cause of female infertility worldwide [2, 3]. Although research on the pathophysiology of granulosa cells (GCs) and oocytes in patients with PCOS has emerged continuously in recent years, critical issues regarding abnormal follicular development in PCOS, such as excessive follicular recruitment, obstruction of dominant follicular selection, and follicular atresia, are still unclear [4, 5]. Human folliculogenesis is a precisely controlled biological process that depends on the synchronization between oocyte maturation and the proliferation of surrounding GCs. The bidirectional communication between oocytes and GCs through signal transduction or direct cell-to-cell contact provides material and structural basis for oocyte-GCs crosstalk, which is necessary for normal follicular development [6]. Besides, oocytes rely on GCs, which provide oocytes with the nutrients and regulatory signals needed to promote oocyte maturity and thus the acquisition of developmental competence [7]. It is poorly understood, however, how changes in GC of PCOS affect follicular development and the crosstalk between oocytes and GCs.
To date, due to the limited availability of GCs and follicles in PCOS patients, most data on changes in GCs affecting follicular development are obtained from animal models. However, human follicles have unique developmental patterns and regulatory networks that differ from animals [8]. Characterizing the changes in the transcriptome in GCs of PCOS and their relationship with follicular development is the key to understand the ovulation disorders in PCOS patients, which in turn is expected to provide opportunities for the development of new diagnostic and therapeutic approaches for improving the fertility of women with PCOS. Studies have revealed the genetic changes of human follicles at different developmental stages and the interaction between oocytes and GCs under physiological conditions [9]. However, the pathogenesis of ovulation dysfunction in patients with PCOS has not been elucidated.
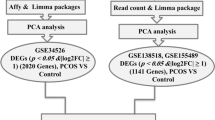
In the current study, we investigated the transcriptome changes in GCs of PCOS from the perspective of follicular development. The flow chart of the search, selection, and analysis of datasets was shown in Scheme 1. The results showed that HAS2 and CBLN1 might be potential therapeutic biomarkers for ovulation disorders in PCOS, and Afamin and TWEAK might be the potential diagnostic and predictive biological markers. Our study provided a reliable research basis for understanding ovulation dysfunction in PCOS and developing targeted interventions aimed to elevate the fertility of PCOS.
Materials and Methods
Microarray Data and Identification of Differentially Expressed Genes
The gene expression profiles of GCs in PCOS were searched in the GEO database (https://www.ncbi.nlm.nih.gov/gds/). The entry type was restricted to “Series,” and the organism was filtered by Homo sapiens. After screening as shown in Scheme 1, GSE34526 was downloaded from the GEO database. The dataset of GSE34526 determined the expression profiling of the GCs from PCOS and normal patients. Besides, the dataset of GSE107746 which identified the transcriptome profiles of normal oocytes and GCs from five follicle development stages was downloaded. The platforms for GSE34526 and GSE107746 were the GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array and the GPL20795 HiSeq X Ten (Homo sapiens), respectively.
Differentially expressed genes (DEGs) with the threshold criterion of p value < 0.05 and fold change (FC) of log2 > 2 in GCs from normal and PCOS patients were screened out with the R limma package (https://www.r-project.org) of the R software program (version 3.5.2). The heatmaps and clustering of DEGs were plotted by the R ggplot2 package in the R analysis platform.
Protein-Protein Interaction
The Search Tool for the Retrieval of Interacting Genes (STRING, https://string-db.org/) database, which is a tool to identify interactions between known genes and predicted genes at the protein level, was used to build the protein-protein interaction (PPI) network specific to DEGs. Cytoscape software (version 3.7.0, https://cytoscape.org/) was employed for visualizing the PPI network. Subsequently, MCODE, a plug-in of Cytoscape, was applied to explore the clusters (highly interconnected regions) in the PPI network.
Gene Ontology Analysis and GSEA Analysis
Gene ontology (GO) analysis was used extensively to identify the characteristic biological attributes of genes, gene products, and sequences, including biological process (BP), cell components (CC), and molecular function (MF). In the current study, GO analysis of DEGs was performed using Metascape (http://metascape.org/gp/index.html) [10]. The Gene Set Enrichment Analysis (GSEA, http://software.broadinstitute.org/gsea/index.jsp) was carried out for all genes with the GSEA software (version 4.0.0) [11], and the gene sets that showed nominal p value less than 0.05 were chosen as enriched.
Genes Related to Oocyte Competence and Follicular Development
Obtained genes which expressed in granulosa cells related to oocyte competence and follicular development through literature search and the Venn diagram tool (bioinformatics.psb.ugent.be/webtools/Venn/) helped identify the intersecting part between the genes and DEGs. Furthermore, the expression level of the DEGs, which are related to oocyte competence and follicular development, in different follicle development stages was got in the dataset of GSE107746.
Genes Encoding Transcription Factors
Animal Transcription Factors Database (AnimalTFDB, http://bioinfo.life.hust.edu.cn/AnimalTFDB/#!/) is a comprehensive database including classification and annotation of genome-wide transcription factors (TFs), transcription co-factors, and chromatin remodeling factors. A total of 1665 human TFs were downloaded from AnimalTFDB [12]. Subsequently, the genes encoding human TFs took the intersection with DEGs by the Venn diagram tool to acquire various TFs involved in the development of PCOS.
Genes Encoding Secretory Protein
GCs-derived secretory proteins may be candidate biomarkers for the diagnosis of PCOS. The Human Protein Atlas (https://www.proteinatlas.org/) maps the human proteins in cells, tissues, and organs using integration of various omics technologies, including antibody-based imaging, mass spectrometry-based proteomics, transcriptomics, and systems biology. All the data in the knowledge resource is open access. The secretory protein-encoding genes were downloaded from the Human Protein Atlas database, and the intersection of DEGs with secreted protein-encoding genes was obtained with the Venn diagram tool.
Statistical Analysis
Statistical analysis was performed using version 3.5.2 of the R software unless otherwise stated. Moreover, all expression data have been normalized to log2. p < 0.05 was taken as a statistically significant difference for all analyses.
Results
DEGs in GCs Between Normal and PCOS Patients
The gene expression profile GSE34526 which composed of 7 PCOS and 3 normal GCs samples was obtained from the GEO database. According to the criterion of p value < 0.05 and log2|FC| > 2, 423 DEGs between normal and PCOS GCs were identified with the R limma package in R analysis platform, including 347 downregulated and 76 upregulated genes (Table S1). Additionally, the relative expression levels of these DEGs were shown in the cluster heatmaps and the volcano plots (Fig. 1). These DEGs could well distinguish the PCOS and normal group completely.
Hierarchical clustering heatmaps and volcanic plots of the differentially expressed genes (DEGs). a Heatmaps of DEGs. Red shading indicates that the expression of genes is relatively upregulated, while blue shading indicates that the expression of genes is relatively downregulated. b Volcanic plots of DEGs. The blue points represent genes with significantly different expression, and the red points represent genes with no significant difference. DEGs were screened on the criterion of log2|FC| > 2.0 and p < 0.05
Key Pathways Involved in PCOS
The GO analysis of the DEGs was established, and the GO analysis was divided into the MF, CC, and BP categories. As shown in Fig. 2, the results revealed that the DEGs were mainly involved in immune response, such as macrophage activation and regulation of cell activation (p < 0.01). Besides, we obtained a PPI network by using the STRING (Fig. 3a). One key module (score = 18.818) was extracted for later analysis (Fig. 3b). For convenience, the module was named as C1QA, containing 23 nodes and 207 edges, of which C1QA, C1QB, FCGR3A, FCGR1A, and CD14 were significant nodes due to the high degree of interconnection. Consequently, the genes that occupy the center of the C1QA module were responsible for immune function.
Functional enrichment analyses of DEGs in GCs of PCOS. a Top 10 molecular functions of EDGs enriched in; b top 10 cellular components of EDGs enriched in; c top 10 biological processes of EDGs enriched in; d top 20 of GO analysis of DEGs. Colored by p value. DEGs, differentially expressed genes; GO, Gene ontology; GCs, granulosa cells
The GSEA analyses were carried out to further elucidate the potential molecular mechanisms and signaling pathways of PCOS (Table S2). Notably, the functional pathways, such as Toll-like receptor, NOTCH, and NOD-like receptor signaling pathways, were significantly representative in GCs of PCOS (p < 0.05) (Fig. 4). Among those pathways, SPP1, PSENEN, NLRC4, and NLRP3 genes were considerably upregulated in the GCs of PCOS. These signaling pathways and upregulated genes most likely mediate the incidence and development of PCOS.
DEGs Related to Oocyte Competence and Follicular Development
Follicular development depends on the synchronization of oocyte maturation and GCs proliferation. At the same time, the maturation of oocytes relies on the nutrients and signaling factors provided by GCs [13]. To clarify whether changes in the transcriptome in GCs of PCOS can affect oocyte quality and follicular development, those genes that have been reported to be expressed in GCs and related to oocyte competence and follicular development inspired our broad interests [9, 14, 15]. Interestingly, we found that CBLN1, which is expressed explicitly in preovulatory GCs, was markedly decreased in GCs of PCOS (Fig. 5a). In addition, HAS2, taking charge of embryo quality and oocyte fertilization rates [16], was visibly downregulated in GCs of PCOS (Fig. 5c).
DEGs related to oocyte competence and follicular development a–d. CBLN1 and HAS2, which should be overexpressed in preovulatory phase, were downregulated in GCs of PCOS. e–f CYP27A1, which should be underexpressed in preovulatory phase, were upregulated in GCs of PCOS. DEGs, differentially expressed genes; GCs, granulosa cells. *: p < 0.05
The GSE107746 dataset from the GEO database was downloaded to investigate the dynamic expression of HAS2 and CBLN1 in GCs during different follicular development stages. The results suggested that HAS2 and CBLN1 expression gradually increased with follicular development and reached a peak in the preovulatory period (Fig. 5b, d). This indicated that HAS2 and CBLN1 might serve as the essential genes triggering ovulation, and the repression of CBLN1 and HAS2 in GCs may function as critical factors for the decline of oocyte quality and follicular development disorders in PCOS.
Furthermore, PCOS is characterized as hyperandrogenemia, and studies have shown that high concentrations of androgen can result in stagnant follicular development [17,18,19]. The close cooperation of theca cells and GCs in the ovary is an integral part of converting androgens into estrogen [20]. We analyzed DEGs related to androgenic biosynthesis and steroid metabolism. Intriguingly, CYP27A1, which should be suppressed in the preovulatory phase, was elevated in GCs of PCOS (Fig. 5e–f), implying that high expression of CYP27A1 in GCs may also exhibit several vital roles in the hyperandrogenemia and follicular atresia in PCOS.
TFs Regulatory Networks in GCs
A total of 1665 known human TFs were downloaded from Animal Transcription Factor Database (TFDB v3.0) [12]. Compared with normal GCs, there are a total of 16 TFs differentially expressed in GCs of PCOS (Fig. 6a). The expressions of MAFB, ASCL3, ASCL2, ZNF620, ZNF154, ZNF296, MNX1, SPI1, CIZ1, ID1, and CIC were upregulated in GCs of PCOS. Conversely, the expressions of ZNF280C, FOXR1, FOXI1, TLX3, and TULP1 were downregulated in GCs of PCOS (Fig. 6b). These genes likely participate in the occurrence and the dysfunction of PCOS.
The immune disorders and apoptosis accumulation of granulosa cells may be the fundamental mechanisms in the pathogenesis of PCOS [21, 22]. PU.1, a TF encode by SPI1, is a transcriptional activator that may be specifically involved in the differentiation or activation of macrophages or B cells [23]. In addition, the ID1 protein is a negatively transcriptional regulator for the basic helix-loop-helix TFs by forming heterodimers and inhibiting their DNA binding and transcriptional activity. It finally regrets a variety of cellular processes, including cellular growth, senescence, differentiation, apoptosis, angiogenesis, and neoplastic transformation [24]. These results manifested that the DEGs which encode TFs may lead to the occurrence and development of PCOS. Further evaluation of these TFs, the upstream signaling pathways, ligands, receptors, and downstream targets in GCs will provide insight into the transcriptional control of PCOS.
Potential Biological Indicators for PCOS
In total, we screened out 14 secretory protein-coding DEGs between normal and PCOS GCs (Fig. 7a). Compared with normal GCs, the expression of AFM and GCG were decreased, and the expression of TNFSF12, SPP1, GRN, ITGAM, SERPINA1, CCL8, FAM20A, FAM20C, PLAU, IL10, CCL2, and TGFB1 were increased in GCs of PCOS (Fig. 7b).
Proteins encoded by SPP1, GRN, ITGAM, CCL8, IL10, and CCL2 participate in immune response [25, 26]. AFM encoded protein (Afamin) functions as a carrier for hydrophobic molecules in body fluids, which is essential for the transport and activity of Wnt family members, such as WNT1, WNT3, WNT5A, WNT7A, and WNT10B [27]. Generally, the Wnt signaling pathway serves a vital role in the apoptosis of GCs [28, 29]. Hence, it illustrated that the decline of Afamin might be a key factor for PCOS development. Moreover, TWEAK protein (encode by TNFSF12) is a weak inducer of apoptosis [30], which may promote the apoptosis of GCs in PCOS. Further evaluation of their expression levels in the biological fluids may contribute to the development of novel biomarkers for PCOS prediction in clinical application.
Discussion
PCOS is a multifactorial disease, and it is well described that there are no worldwide treatments for PCOS [31]. The current study revealed the transcriptome changes in GCs of PCOS from the perspective of follicular development. DEGs and signaling pathways which may be involved in the occurrence of PCOS were analyzed. In addition, we found several secretory proteins that may work as potential biomarkers. What is more, we found several DEGs which may cause oocyte competence depletion and follicular atresia. These findings may provide valuable clues for future studies in the diagnosis and treatment of PCOS.
This study proposed a set of potentially secreted proteins that can be used as biological indicators. Most of the secretory proteins were likely involved in immune responses of PCOS patients. Further study of these secreted proteins expression in biological fluids, such as blood and follicular fluid, may be beneficial for the development of new diagnostic and therapeutic approaches to PCOS.
PCOS patients often seek the help of assisted reproduction technology due to ovulation dysfunction [32]. Studies have demonstrated that the oocyte maturation rate of patients with PCOS is decreased in vitro [33]. The bidirectional communication between oocytes and granulosa cells is a pivotal material and structural basis for the maturation of oocytes [34]. In this study, we found that HAS2 and CBLN1 were downregulated in GCs of PCOS. HAS2 and CBLN1 expression should increase with follicular development and reach a peak in the preovulatory period [9], and the expression level of HAS2 and CBLN1 in GCs may affect oocyte quality. Further research on these factors may help reveal the pathogenesis of low oocyte maturation rate and follicular atresia in patients with PCOS.
PCOS is often accompanied by insulin resistance and hyperinsulinemia [35]. We firstly found that MAFB, a TF activating the insulin and glucagon promoters [36], was upregulated in GCs of PCOS. MAFB is required for islet beta-cell differentiation and can weakly transactivate the glucagon gene promoter through the G1 element together with PAX6 [37]. In addition, we found that GCG, which encodes glucagon, was downregulated in GCs of PCOS. Glucagon plays a crucial role in glucose metabolism and homeostasis [38], illustrating that the reduction of GCG may lead to the disorders of blood glucose metabolism in PCOS. In conclusion, we found several factors, which may be vitally involved in the formation of insulin resistance of PCOS. Further study of these factors may facilitate to clarify the etiology of insulin resistance in PCOS patients.
In summary, the current study found several factors, such as Afamin and TWEAK, which may be the potential diagnostic and predictive biological markers, and some signaling pathway, for example, Toll-like receptor, NOTCH, and NOD-like receptor signaling pathways, which may participate in the occurrence and development of PCOS. This study also found several DEGs, including HAS2 and CBLN1 that were related to the oocyte competence and follicle development. This work provided a valuable resource for developing targeted interventions aimed to elevate the fertility of PCOS.
References
Risal S, Pei Y, Lu H, Manti M, Fornes R, Pui HP, et al. Prenatal androgen exposure and transgenerational susceptibility to polycystic ovary syndrome. Nat Med. 2019;25(12):1894–904. https://doi.org/10.1038/s41591-019-0666-1.
Jayasena CN, Franks S. The management of patients with polycystic ovary syndrome. Nat Rev Endocrinol. 2014;10(10):624–36. https://doi.org/10.1038/nrendo.2014.102.
Tata B, Mimouni NEH, Barbotin AL, Malone SA, Loyens A, Pigny P, et al. Elevated prenatal anti-Mullerian hormone reprograms the fetus and induces polycystic ovary syndrome in adulthood. Nat Med. 2018;24(6):834–46. https://doi.org/10.1038/s41591-018-0035-5.
Jonard S, Dewailly D. The follicular excess in polycystic ovaries, due to intra-ovarian hyperandrogenism, may be the main culprit for the follicular arrest. Hum Reprod Update. 2004;10(2):107–17. https://doi.org/10.1093/humupd/dmh010.
Franks S, Stark J, Hardy K. Follicle dynamics and anovulation in polycystic ovary syndrome. Hum Reprod Update. 2008;14(4):367–78. https://doi.org/10.1093/humupd/dmn015.
Chang HM, Qiao J, Leung PC. Oocyte-somatic cell interactions in the human ovary-novel role of bone morphogenetic proteins and growth differentiation factors. Hum Reprod Update. 2016;23(1):1–18. https://doi.org/10.1093/humupd/dmw039.
Sanchez F, Smitz J. Molecular control of oogenesis. Biochim Biophys Acta. 2012;1822(12):1896–912. https://doi.org/10.1016/j.bbadis.2012.05.013.
Knight PG, Glister C. TGF-beta superfamily members and ovarian follicle development. Reproduction. 2006;132(2):191–206. https://doi.org/10.1530/rep.1.01074.
Zhang Y, Yan Z, Qin Q, Nisenblat V, Chang HM, Yu Y, et al. Transcriptome landscape of human folliculogenesis reveals oocyte and granulosa cell interactions. Mol Cell. 2018;72(6):1021–34 e4. https://doi.org/10.1016/j.molcel.2018.10.029.
Niu X, Zhang J, Zhang L, Hou Y, Pu S, Chu A, et al. Weighted gene co-expression network analysis identifies critical genes in the development of heart failure after acute myocardial infarction. Front Genet. 2019;10:1214. https://doi.org/10.3389/fgene.2019.01214.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. https://doi.org/10.1073/pnas.0506580102.
Zhang HM, Chen H, Liu W, Liu H, Gong J, Wang H, et al. AnimalTFDB: a comprehensive animal transcription factor database. Nucleic Acids Res. 2012;40(Database issue):D144–9. https://doi.org/10.1093/nar/gkr965.
Ford E, Beckett EL, Roman S, McLaughlin EA, Sutherland J. Advances in human primordial follicle activation and premature ovarian insufficiency. Reproduction. 2019;159:R15–29. https://doi.org/10.1530/REP-19-0201.
Huang Z, Wells D. The human oocyte and cumulus cells relationship: new insights from the cumulus cell transcriptome. Mol Hum Reprod. 2010;16(10):715–25. https://doi.org/10.1093/molehr/gaq031.
Parks JC, Patton AL, McCallie BR, Griffin DK, Schoolcraft WB, Katz-Jaffe MG. Corona cell RNA sequencing from individual oocytes revealed transcripts and pathways linked to euploid oocyte competence and live birth. Reprod BioMed Online. 2016;32(5):518–26. https://doi.org/10.1016/j.rbmo.2016.02.002.
Bhardwaj R, Ansari MM, Pandey S, Parmar MS, Chandra V, Kumar GS, et al. GREM1, EGFR, and HAS2; the oocyte competence markers for improved buffalo embryo production in vitro. Theriogenology. 2016;86(8):2004–11. https://doi.org/10.1016/j.theriogenology.2016.06.019.
Summers AF, Pohlmeier WE, Sargent KM, Cole BD, Vinton RJ, Kurz SG, et al. Altered theca and cumulus oocyte complex gene expression, follicular arrest and reduced fertility in cows with dominant follicle follicular fluid androgen excess. PLoS One. 2014;9(10):e110683. https://doi.org/10.1371/journal.pone.0110683.
Chou CH, Chen MJ. The effect of steroid hormones on ovarian follicle development. Vitam Horm. 2018;107:155–75. https://doi.org/10.1016/bs.vh.2018.01.013.
Rosenfield RL, Ehrmann DA. The pathogenesis of polycystic ovary syndrome (PCOS): the hypothesis of PCOS as functional ovarian hyperandrogenism revisited. Endocr Rev. 2016;37(5):467–520. https://doi.org/10.1210/er.2015-1104.
Dewailly D, Robin G, Peigne M, Decanter C, Pigny P, Catteau-Jonard S. Interactions between androgens, FSH, anti-Mullerian hormone and estradiol during folliculogenesis in the human normal and polycystic ovary. Hum Reprod Update. 2016;22(6):709–24. https://doi.org/10.1093/humupd/dmw027.
Gong P, Shi B, Wang J, Cao P, Diao Z, Wang Y, et al. Association between Th1/Th2 immune imbalance and obesity in women with or without polycystic ovary syndrome. Gynecol Endocrinol. 2018;34(8):709–14. https://doi.org/10.1080/09513590.2018.1428301.
Jiang B, Xue M, Xu D, Song J, Zhu S. Down-regulated lncRNA HOTAIR alleviates polycystic ovaries syndrome in rats by reducing expression of insulin-like growth factor 1 via microRNA-130a. J Cell Mol Med. 2020;24(1):451–64. https://doi.org/10.1111/jcmm.14753.
Fischer J, Walter C, Tonges A, Aleth H, Jordao MJC, Leddin M, et al. Safeguard function of PU.1 shapes the inflammatory epigenome of neutrophils. Nat Immunol. 2019;20(5):546–58. https://doi.org/10.1038/s41590-019-0343-z.
Sun B, Dong C, Lei H, Gong Y, Li M, Zhang Y, et al. Knockdown of inhibitor of differentiation 1 suppresses proliferation and induces apoptosis by inactivating PI3K/Akt/mTOR signaling in hemangioma-derived endothelial cells. Biomed Pharmacother. 2019;111:236–43. https://doi.org/10.1016/j.biopha.2018.12.072.
DiScipio RG, Daffern PJ, Schraufstatter IU, Sriramarao P. Human polymorphonuclear leukocytes adhere to complement factor H through an interaction that involves alphaMbeta2 (CD11b/CD18). J Immunol. 1998;160(8):4057–66.
Beel S, Moisse M, Damme M, De Muynck L, Robberecht W, Van Den Bosch L, et al. Progranulin functions as a cathepsin D chaperone to stimulate axonal outgrowth in vivo. Hum Mol Genet. 2017;26(15):2850–63. https://doi.org/10.1093/hmg/ddx162.
Mihara E, Hirai H, Yamamoto H, Tamura-Kawakami K, Matano M, Kikuchi A, et al. Active and water-soluble form of lipidated Wnt protein is maintained by a serum glycoprotein afamin/alpha-albumin. Elife. 2016;5. https://doi.org/10.7554/eLife.11621.
Mehdinejadiani S, Amidi F, Mehdizadeh M, Barati M, Safdarian L, Aflatoonian R, et al. The effects of letrozole and clomiphene citrate on ligands expression of Wnt3, Wnt7a, and Wnt8b in proliferative endometrium of women with polycystic ovarian syndrome. Gynecol Endocrinol. 2018;34(9):775–80. https://doi.org/10.1080/09513590.2018.1446934.
Jiang YC, Ma JX. The role of MiR-324-3p in polycystic ovary syndrome (PCOS) via targeting WNT2B. Eur Rev Med Pharmacol Sci. 2018;22(11):3286–93. https://doi.org/10.26355/eurrev_201806_15147.
Zimmermann M, Koreck A, Meyer N, Basinski T, Meiler F, Simone B, et al. TNF-like weak inducer of apoptosis (TWEAK) and TNF-alpha cooperate in the induction of keratinocyte apoptosis. J Allergy Clin Immunol. 2011;127(1):200–7, 7 e1–10. https://doi.org/10.1016/j.jaci.2010.11.005.
Nejabati HR, Samadi N, Roshangar L, Nouri M. N1-methylnicotinamide as a possible modulator of cardiovascular risk markers in polycystic ovary syndrome. Life Sci. 2019;235:116843. https://doi.org/10.1016/j.lfs.2019.116843.
Palomba S, Daolio J, La Sala GB. Oocyte competence in women with polycystic ovary syndrome. Trends Endocrinol Metab. 2017;28(3):186–98. https://doi.org/10.1016/j.tem.2016.11.008.
Qiao J, Feng HL. Extra- and intra-ovarian factors in polycystic ovary syndrome: impact on oocyte maturation and embryo developmental competence. Hum Reprod Update. 2011;17(1):17–33. https://doi.org/10.1093/humupd/dmq032.
Guo J, Zhang T, Guo Y, Sun T, Li H, Zhang X, et al. Oocyte stage-specific effects of MTOR determine granulosa cell fate and oocyte quality in mice. Proc Natl Acad Sci U S A. 2018;115(23):E5326–E33. https://doi.org/10.1073/pnas.1800352115.
Azziz R, Carmina E, Chen Z, Dunaif A, Laven JS, Legro RS, et al. Polycystic ovary syndrome. Nat Rev Dis Primers. 2016;2:16057. https://doi.org/10.1038/nrdp.2016.57.
Bethea M, Liu Y, Wade AK, Mullen R, Gupta R, Gelfanov V, et al. The islet-expressed Lhx1 transcription factor interacts with Islet-1 and contributes to glucose homeostasis. Am J Physiol Endocrinol Metab. 2019;316(3):E397–409. https://doi.org/10.1152/ajpendo.00235.2018.
Xiafukaiti G, Maimaiti S, Ogata K, Kuno A, Kudo T, Shawki HH et al. MafB is important for pancreatic beta-cell maintenance under a MafA-deficient condition. Mol Cell Biol. 2019;39(17). doi:https://doi.org/10.1128/MCB.00080-19.
Billing LJ, Larraufie P, Lewis J, Leiter A, Li J, Lam B, et al. Single cell transcriptomic profiling of large intestinal enteroendocrine cells in mice - identification of selective stimuli for insulin-like peptide-5 and glucagon-like peptide-1 co-expressing cells. Mol Metab. 2019;29:158–69. https://doi.org/10.1016/j.molmet.2019.09.001.
Availability of Data and Material
The data underlying this article are available in the article and in its online supplementary material.
Funding
This work was supported by the National Key R&D Program of China (2018YFC1003200) and National Natural Science Foundation of China (81860276, 11774274).
Author information
Authors and Affiliations
Contributions
Dongyong Yang and Yanxiang Cheng made the design of this study. Na Li, Aiping Ma, Fangfang Dai, and Yajing Zheng did the acquisition and analysis of data. Xuejia Hu, Yanqing Wang, Shu Xian, Mengqin Yuan, and Shiyi Liu did the interpretation of data. Dongyong Yang and Zhimin Deng drafted the article. Yi Yang and Yanxiang Cheng revised this study critically. All authors approved of the version to be published.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interest.
Declarations
Not applicable.
Code Availability
All code used during the study are available from the corresponding author by request.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Dongyong Yang, Na Li, and Aiping Ma are authors should be regarded as joint First Authors.
Rights and permissions
About this article
Cite this article
Yang, D., Li, N., Ma, A. et al. Identification of Potential Biomarkers of Polycystic Ovary Syndrome via Integrated Bioinformatics Analysis. Reprod. Sci. 28, 1353–1361 (2021). https://doi.org/10.1007/s43032-020-00352-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43032-020-00352-x