Abstract
The aim of this study was to isolate Aspergillus section Nigri from onion samples bought in supermarkets and to analyze the fungal isolates by means of molecular data in order to differentiate A. niger and A. welwitschiae species from the other non-toxigenic species of black aspergilli, and detect genes involved in the biosynthesis of ochratoxin A and fumonisin B2. Aspergillus section Nigri were found in 98% (94/96) of the onion samples. Based on the results of multiplex PCR (performed on 500 randomly selected strains), 97.4% of the Aspergillus section Nigri strains were recognized as A. niger/A. welwitschiae. Around half of them were subjected to partial sequencing of the CaM gene to distinguish one from the other. A total of 97.9% of the isolates were identified as A. welwitschiae and only 2.1% as A. niger. The fum8 gene, involved in fumonisin B2 biosynthesis, was found in 36% of A. welwitschiae isolates, but radH and pks genes, involved in ochratoxin A biosynthesis, were found in only 2.8%. The presence/absence of fum8 gene in the A. welwitschiae genome is closely associated with ability/inability of the isolates to produce fumonisin in vitro. Based on these results, we suggest that in-depth studies are conducted to investigate the presence of fumonisins in onion bulbs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
It is not easy to recognize the macro- and micro-morphological differences between some species of Aspergillus section Nigri (black aspergilli). The use of molecular methods has proved essential for distinguishing these species. Nowadays, 27 species are included in A. section Nigri which, based on calmodulin gene sequences, is split into seven clades: A. tubingensis, A. niger, A. brasiliensis, A. carbonarius, A. heteromorphus, A. homomorphus, and A. aculeatus [1, 2].
Ten morphologically very similar taxa, A. costaricaensis, A. luchuensis, A. neoniger, A. piperis, A. tubingensis, A. eucalypticola and A. vadensis (A. tubingensis clade), A. niger and A. welwitschiae (A. niger clade), and A. brasiliensis (A. brasiliensis clade), are generally referred as Aspergillus niger aggregate [2]. The significance of this group of fungi as contaminants of food products regarding their occurrence and their ability to produce mycotoxins has been investigated worldwide. Two mycotoxins commonly associated with Aspergillus niger aggregate are fumonisin B2 (FB2) and ochratoxin A (OTA). However, only fungal strains belonging to A. niger and A. welwitschiae have been reported to be able to produce these mycotoxins in culture media as well as on natural substrates [3,4,5]. A. welwitschiae is a species which, not too long ago, was dismembered from the A. niger taxon [3, 6]. No other species belonging to A. niger aggregate have been described as OTA and/or FB2 producers. OTA is a nephrotoxic and carcinogenic mycotoxin and FB2 is neurotoxic and hepatotoxic to animal species, and has also been associated with esophageal cancers in humans [7, 8].
Considering the inherent risk of the occurrence of A. niger and A. welwitschiae in foodstuffs, several molecular methods have been developed for distinguishing these toxigenic species from non-toxigenic A. niger aggregate species, including PCR using the primer-pair benA-An/Aw designed by Massi et al. [9]. Using this primer-pair, only those PCRs containing A. niger or A. welwitschiae template DNA produce an amplicon of 192 bp in length. Because not all strains of A. niger and A. welwitschiae are able to produce OTA or FB2, some authors have investigated the presence of some essential genes involved in OTA or FB2 synthesis in A. niger and A. welwitschiae strains isolated from foodstuffs, with the aim of predicting the contamination risk [9, 10]. It is consensual that the fum8 (α-oxoamine synthase) and the pks An15g07920 (polyketide synthase) genes are required respectively for the production of FB2 and OTA by Aspergillus [9,10,11]. Based on this confirmation, Massi et al. [9] developed a multiplex PCR technique to detect the frequency of A. niger/A. welwitschiae strains harboring these genes essential for the production of OTA and FB2.
A. niger aggregate species are frequently responsible for post-harvest losses of some vegetables. Black mold is one of the predominant post-harvest onion diseases, and was initially thought to be caused by A. niger [12]. However, there are very few molecular biology–based studies to distinguish A. niger and A. welwitschiae from other A. niger aggregate species isolated from onion samples [9, 13, 14]. Gherbawy et al. [14] detected black aspergilli in 92.5% of onion samples produced in the Taif region of Saudi Arabia. Using molecular data, the authors showed that the most common fungus isolated from the onion bulb was A. welwitschiae. Of 37 isolates of A. welwitschiae analyzed by Gherbawy et al. [14], 18 isolates were fumonisin producers, while none of them was able to produce ochratoxins.
Given that onions are one of the most widely consumed fresh vegetables in Brazil, the aim of this study was to isolate Aspergillus section Nigri from onion bulb samples, bought in supermarkets over a period of 24 consecutive months, and to analyze the fungal isolates by means of molecular data in order to differentiate A. niger and A. welwitschiae species from the other non-toxigenic species of black aspergilli, and detect genes involved in the biosynthesis of OTA and FB2.
Materials and methods
Isolation of fungi from onion samples
The yellow variety of onions (Allium cepa L.) sold in supermarkets in Paraná State, southern Brazil, was analyzed for the presence of fungi. A total of ninety-six samples purchased over 24 consecutive months in Paraná State, Brazil, were collected from four different supermarkets. Approximately 0.3 kg of onion samples, apparently with no ongoing disease, was selected randomly from the supermarket gondolas, placed in plastic bags and taken to the laboratory.
The dry outer and fleshy inner scales of the onions were cut into pieces of 1 × 1 cm and disinfected superficially by immersion in 0.4% sodium hypochlorite solution (NaOCl) for 2 min [15]. The excess moisture was removed by blotting on sterile filter paper. One hundred pieces (dry and fleshy) from the bulb were plated on Dichloran 18% Glycerol agar (DG18 agar) [16]. A portion containing adventitious roots was also disinfected and plated. The plates were incubated at 25 °C for 5–7 days. The incidence of fungal genera was determined morphologically [2, 15, 17]. Colonies belonging to Aspergillus section Nigri were isolated, purified on Czapek Yeast Autolysate agar (CYA agar - [Pitt 1979]) and incubated at 25 °C for 7 days.
DNA extraction from Aspergillus section Nigri
Five hundred Aspergillus section Nigri isolates were grown in 7 mL of liquid complete medium [18] at 25 °C for 24 h. The mycelia were collected, frozen in liquid nitrogen, and ground to a fine powder. Genomic DNA was extracted using the BioPur Mini Spin Extraction Kit® (Biometrix, Brazil), according to the manufacturer’s instructions.
Multiplex PCR
Five hundred Aspergillus section Nigri isolates were analyzed using the multiplex PCR method described by Massi et al. [9] in order to differentiate A. niger/A. welwitschiae species from the remainder of A. section Nigri species and also to characterize the strains in terms of the presence or absence of the two genes required for ochratoxin biosynthesis (radH gene, locus tag An15g07880 of A. niger CBS 513.88, and the pks gene, locus tag An15g07920 of A. niger CBS 513.88), respectively, encoding a flavin-dependent halogenase and a polyketide synthase, and the gene required for FB2 synthesis (fum8 gene, locus tag An01g06870 of A. niger CBS 513.88) encoding an α-oxoamine synthase.
Partial calmodulin gene sequence analysis
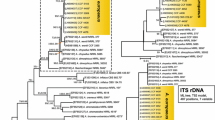
Two hundred fifty Aspergillus section Nigri isolates were subjected to partial sequencing of the calmodulin (CaM) gene. The CaM amplicon obtained using the primer-pair cmd5/cmd6, according to the protocol described by Hong et al. [19], was purified using ExoProStar™ 1-Step (GE Healthcare Life Sciences, UK). The purified amplicon was sequenced using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA) in an ABI3500XL genetic analyzer (Applied Biosystems, USA). The sequences obtained were aligned with sequences from all Aspergillus section Nigri type strains in the NCBI database (http://www.ncbi.nlm.nih.gov/) using ClustalW and BioEdit software [20]. Phylogenetic analysis was run using MEGA7 software [21] and the maximum likelihood (ML) method with the best model. To determine the support for each clade, bootstrap analysis was run on 1000 replicates.
FB2 production by A. welwitschiae strains
A total of 50 A. welwitschiae strains (24 PCR-positive for fum8 and 26 PCR-negative for fum8) were analyzed for FB2 production. Briefly, the strains were inoculated onto Czapek Yeast Extract 20% Sucrose agar (CY20S agar). The plates were incubated for 7 days at 25 °C. The toxin was extracted from five small plugs of mycelium with methanol. The extract was filtered and FB2 derivatized using ortho-phthaldialdehyde reagent (OPA) and then injected into a Shimadzu LC-10VP (Shimadzu, Japan) HPLC system, with a fluorescence detector set to 335-nm excitation and 440-nm emission. The chromatography column used was a YMC-Pack ODS-A (YMC Co., Japan) (5 mm, 4.6 × 150 mm) with a mobile phase of acetonitrile:water:acetic acid (51:47:02, v/v/v). The respective flow rate, oven temperature, and injection volume were 1 mL/1 min, 40 °C, and 20 μL. The methodology is described in detail in Ferranti et al. [22].
Results
All outer dry scales, inner fleshy scales, and all adventitious root samples analyzed herein showed fungal infection. Based on macroscopic and microscopic examinations, we identified Penicillium (51.1%) and Aspergillus (43.4%) as the predominant fungal genera in all onion samples. Other fungal genera, such as Fusarium and Cladosporium, were observed at low frequency (5.4%). Among the Aspergillus counted in the onion samples, those belonging to the Aspergillus section Nigri were the most frequent (99.8%) and found in 98% (94/96) of the onion samples. The highest incidence of Aspergillus section Nigri was found in the inner scale (50.2%), where water activity values corroborate with the highest number of fungal contaminations, followed by outer dry scales (41.5%) and adventitious roots (8.3%). The frequency of occurrence and average of infection by Aspergillus section Nigri species are given in Table 1.
Based on the results of multiplex PCR (performed on 500 strains randomly selected), 97.4% of the Aspergillus section Nigri strains (n = 487) were recognized as A. niger/A. welwitschiae. Only 13 strains (2.6%) did not reveal the predicted amplicon when using the specific primers developed for detecting A. niger/A. welwitschiae.
The Aspergillus section Nigri strains (n = 13) that did not reveal any multiplex PCR product were subjected to partial sequencing of the calmodulin (CaM) gene for identification purposes. Four strains were phylogenetically identified as A. brasiliensis, three as A. japonicus, two as A. tubingensis, two as A. luchuensis, one as A. neoniger, and one as A. heteromorphus (Fig. 1).
Of the 487 strains recognized as A. niger/A. welwitschiae, approximately half (n = 242) were also subjected to partial sequencing of the CaM gene to distinguish one from the other. Based on the percentage identity found using BLASTn and a phylogenetic inference (data not shown), 237 strains (97.9%) were identified as A. welwitschiae and only five (2.1%) as A. niger.
The gene fum8, essential for FB2 synthesis, was found in 36% of A. welwitschiae isolates, whereas the radH and pks genes, involved in OTA biosynthesis, were found in only 2.8% of the isolates. Because the incidence of fum8 was high, we examined FB2 production in a sample of 50 A. welwitschiae strains (24 PCR-positive for fum8 and 26 PCR-negative for fum8). Approximately 96% of the PCR-positive strains produced FB2 in Czapek Yeast Extract 20% Sucrose (CY20S) agar. On the other hand, 100% of PCR-negative strains were FB2 non-producing, meaning that the loss of this capability is closely associated with gene deletions within the A. welwitschiae genome. In the present study, the number of positive strains for OTA genes was low (n = 7) and the association between strains that are positive for the radH and pks genes and their actual production of OTA was not investigated.
Discussion
There are very few studies on the presence of toxigenic fungi in onion bulbs and the potential risk associated with mycotoxin production. In our study, we investigated fungal incidence in onion samples purchased over 24 consecutive months in Paraná State, Brazil. In line with the findings for onion bulbs marketed in Korea [23], Hungary [13], Saudi Arabia [14], Bangladesh [24], and Nigeria [25], Penicillium and Aspergillus were the genera most frequently found.
Aspergillus section Nigri was isolated from 98% of the onion samples. The multiplex PCR allowed A. niger/A. welwitschiae strains to be differentiated from the remainder of Aspergillus section Nigri species. Of the 500 randomly sampled fungal isolates, A. niger/A. welwitschiae were much more frequent than other A. niger aggregate species throughout the period herein investigated. These results are in line with studies that have shown A. niger [25, 26] or A. welwitschiae [9, 13, 14] to be the prevalent species in onion bulbs.
Perrone and coauthors have shown that the CaM sequence can be used to differentiate A. niger and A. welwitschiae [3] and based on this type of gene sequence, some authors have reported that the frequency of occurrence of A. niger and A. welwitschiae can vary in different hosts [27,28,29]. In our study, every month for 24 months, we selected A. niger/A. welwitschiae isolates for partial sequencing of CaM with the aim of differentiating one from the other. Based on the percentage identity provided by BLASTn and a phylogenetic tree (data not shown), 98% of the strains were identified as A. welwitschiae. The number of isolates herein analyzed (n = 250) was substantially greater than that in the previous studies, which allows us to consistently reveal that A. welwitschiae is the most frequent black aspergilli found throughout the year.
The 13 Aspergillus section Nigri isolates that did not result in any PCR product when using A. niger/A. welwitschiae–specific primers were identified as A. brasiliensis (n = 4), A. japonicus (n = 3), A. tubingensis (n = 2), A. luchuensis (n = 2), A. neoniger (n = 1), and A. heteromorphus (n = 1). None of these species is OTA and/or FB2 producers.
In fungal genomes, genes associated with mycotoxin biosynthesis are often clustered, co-regulated, and co-expressed. These gene clusters usually harbor genes encoding one or more types of “core” enzyme responsible for biosynthesis of the metabolite backbone structure [30]. They may contain polyketide synthases (PKS) and non-ribosomal peptide synthetases (NRPS), which are adjacent to genes encoding regulatory proteins, hydrolases, oxidases, methylases, etc., that are involved in chemical transformations of the backbone structure to produce the final metabolite [31]. Ferracin et al. [32] investigated a gene cluster harboring one pks gene, annotated as An15g07920 in the genome of A. niger CBS 513.88, predicted by Pel et al. [33] to be involved in OTA biosynthesis. This gene cluster is located near to the end of chromosome III. In many eukaryotic genomes, chromosome ends are sites of higher rates of rearrangement, such as deletion, compared to the rest of the genome. Since not all the strains of A. niger are able to produce OTA [9, 14, 34], Ferracin et al. [32] investigated 119 A. niger strains for the presence/absence of this pks gene and also for the ability to produce OTA. The pks gene was detected in all strains that produced OTA (n = 31), but not detected in any of the non-producing strains (n = 88), revealing a positive association between the ability to produce OTA and the presence of the pks locus in the A. niger/A. welwitschiae genome. Castellá and Cabanes [35] also showed the importance of the An15g07920 pks gene for specific detection of OTA-producing strains of A. niger aggregate. It is important to mention that the authors’ findings were reported before A. niger sensu stricto was dismembered into two taxa, A. niger and A. welwitschiae [3, 6]. Gherbawy et al. [14] analyzed A. welwitschiae strains (n = 37) isolated from onion samples from Saudi Arabia. The authors found that all strains were non-OTA producing and the pks gene involved in OTA biosynthesis was missing in all of them. In our study, we analyzed a large number of A. welwitschiae strains (n = 237) isolated from onions and showed that the great majority (97.2%) of the A. welwitschiae isolates do not have the two essential genes involved in ochratoxin biosynthesis.
The fumonisin biosynthetic gene cluster in A. niger contains at least 14 fum genes [36]. Surveys of some of these genes in the fumonisin biosynthetic gene (fum) cluster have indicated that fumonisin-non-producing isolates of Aspergillus welwitschiae lack some of them, but non-producing isolates of Aspergillus niger do not [9, 27, 29]. In our study, the use of the multiplex PCR procedure revealed that 36% of the A. welwitschiae strains obtained from onions were positive for the fum8 gene, which encodes an alpha-oxoamine synthase [10]. The presence/absence of this gene in the A. welwitschiae genome is closely associated with ability/inability of the isolates to produce fumonisin in vitro. In line with our findings, Gherbawy et al. [14] reported that 18 of the 37 A. welwitschiae isolates collected from onion samples purchased at markets in Saudi Arabia were potential producers of FB2. These authors also examined the FB2 content in onion samples and detected this mycotoxin in 37.5%. Mycotoxin contamination of Brazilian onion bulbs has not been investigated, but currently we are working to check for the presence of FB2 in several samples of Brazilian onions.
Conclusion
We conclude that A. welwitschiae is very frequently found in onion bulbs purchased at markets. Of the Aspergillus species, A. welwitschiae is the most prevalent throughout the year. Essential genes involved in OTA biosynthesis are missing in almost all A. welwitschiae strains, but the fum8 gene required for fumonisin biosynthesis is present in approximately 36% of the strains. The presence/absence of the fum8 gene in A. welwitschiae strains is closely associated with ability/inability to produce fumonisin in vitro. The presence of fumonisins in Brazilian onion bulbs needs to be investigated in order to clarify the significance of these observations.
References
Fungaro MHP, Ferranti LS, Massi FP, da Silva JJ, Sartori D, Taniwaki MH, Frisvad JC, Iamanaka BT (2017) Aspergillus labruscus sp. nov., a new species of Aspergillus section Nigri discovered in Brazil. Sci Rep 7:6203. https://doi.org/10.1038/s41598-017-06589-y
Samson RA, Visagie CM, Houbraken J, Hong SB, Hubka V, Klaassen CHW, Perrones G, Seifert KA, Susca A, Tanney JB, Varga J, Kocsube S, Szigeti G, Yaguchi T, Frisvad JC (2014) Phylogeny, identification and nomenclature of the genus Aspergillus. Stud Mycol 78:141–173. https://doi.org/10.1016/j.simyco.2014.07.004
Perrone G, Stea G, Epifani F, Varga J, Frisvad JC, Samson RA (2011) Aspergillus niger contains the cryptic phylogenetic species A. awamori. Fungal Biol 115:1138–1150. https://doi.org/10.1016/j.funbio.2011.07.008
Abarca ML, Bragulat MR, Castellá G, Cabañes FJ (1994) Ochratoxin A production by strains of Aspergillus niger var. niger. Appl Environ Microbiol 60:2650–2652. https://doi.org/10.1128/AEM.60.7.2650-2652.1994
Frisvad JJ, Smedsgaard J, Samson RA, Larsen TO, Thrane U (2007) Fumonisin B2 production by Aspergillus niger. J Agric Food Chem 55:9727–9732. https://doi.org/10.1021/jf0718906
Hong SB, Lee M, Kim DH, Varga J, Frisvad JC, Perrone G, Gomi K, Yamada O, Machida M, Houbraken J, Samson RA (2013) Aspergillus luchuensis, an industrially important black Aspergillus in East Asia. PLoS One 8:1–97. https://doi.org/10.1371/journal.pone.0063769
Voss KA, Riley RT (2013) Fumonisin toxicity and mechanism of action: overview and current perspectives. Food Safety 1:2013006. https://doi.org/10.14252/foodsafetyfscj.2013006
Malir F, Ostry V, Pfohl-Leszkowicz A, Malir J, Toman J (2016) Ochratoxin A: 50 years of research. Toxins 8:12–15. https://doi.org/10.3390/toxins8070191
Massi FP, Sartori D, Ferranti LS, Iamanaka BT, Taniwaki MH, Vieira MLC, Fungaro MHP (2016) Prospecting for the incidence of genes involved in ochratoxin and fumonisin biosynthesis in Brazilian strains of Aspergillus niger and Aspergillus welwitschiae. Int J Food Microbiol 221:19–28. https://doi.org/10.1016/j.ijfoodmicro.2016.01.010
Shimizu K, Nakagawa H, Hashimoto R, Hagiwara D, Onji Y, Asano K, Kawamoto S, Takahashi H, Yokoyama K (2015) The a-oxoamine synthase gene fum8 is involved in fumonisin B2 biosynthesis in Aspergillus niger. Mycoscience 56:1–8. https://doi.org/10.1016/j.myc.2014.09.001
Zhang H, Apaliya MT, Mahunu GK, Chen L, Li W (2016) Control of ochratoxin A-producing fungi in grape berry by microbial antagonists: a review. Trends Food Sci Technol 51:88–97. https://doi.org/10.1016/j.tifs.2016.03.012
Walker JC (1924) Further studies on relation of onion scale pigmentation to disease resistance. J Agric Res XXIX:507–514
Varga J, Kocsubé S, Szigeti G, Man V, Tóth B, Vágvölgyi C, Bartók T (2012) Black aspergilli and fumonisin contamination in onions purchased in Hungary. Acta Aliment 41:414–423. https://doi.org/10.1556/AAlim.41.2012.4.3
Gherbawy Y, Elhariry H, Kocsubé S, Bahobial A, El Deeb B, Altalhi A, Varga J, Vágvölgyi C (2015) Molecular characterization of black Aspergillus species from onion and their potential for ochratoxin A and fumonisin B2 production. Foodborne Pathog Dis 12(5):414–413. https://doi.org/10.1089/fpd.2014.1870
Pitt JI, Hocking AD (2009) Fungi and food spoilage, Second edn. Black Academic & Professional - Chapman & Hall, London, 593p. https://doi.org/10.1007/978-0-387-92207-2
Hocking AD, Pitt JI (1980) Dichloran-glycerol medium for enumeration of xerophilic fungi from low moisture foods. Appl Environ Microbiol 39:488–492. https://doi.org/10.1128/AEM.39.3.488-492.1980
Visagie CM, Houbraken J, Frisvad JC, Hong SB, Klaassen CHW, Perrone G, Seifert KA, Varga J, Yaguchi T, Samson RA (2014) Identification and nomenclature of the genus Penicillium. Stud Mycol 78:343–371. https://doi.org/10.1016/j.simyco.2014.09.001
Pontecorvo G, Roper JA, Hemmons LM, MacDonald KD, Buffon AWJ (1953) The genetics of Aspergillus nidulans. Adv Genet 5:141–148. https://doi.org/10.1016/S0065-2660(08)60408-3
Hong SB, Cho HS, Shin HD (2006) Novel Neosartorya species isolated from soil in Korea. Int J Syst Evol Microbiol 56:477–486. https://doi.org/10.1099/ijs.0.63980-0
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Ferranti LS, Fungaro MHP, Massi FP, Silva JJ, Penha RES, Frisvad JC, Taniwaki MH, Iamanaka BT (2018) Diversity of Aspergillus section Nigri on the surface of Vitis labrusca and its hybrid grapes. Int J Food Microbiol 268:53–60. https://doi.org/10.1016/j.ijfoodmicro.2017.12.027
Sang MK, Han GD, Oh JY, Chun SC, Kim KD (2014) Penicillium brasilianum as a novel pathogen of onion (Allium cepa L.) and other fungi predominant on market onion in Korea. Crop Prot 65:138–142. https://doi.org/10.1016/j.cropro.2014.07.016
Ara MAM, Khatun ML, Ashrafuzzaman M (2008) Fungi causing rots in onions at storage and market. J Bangladesh Agril Univ 6:245–251. https://doi.org/10.3329/jbau.v6i2.4818
Samuel O, Ifeanyi O (2015) Fungi associated with the deterioration of post-harvest onion bulbs sold in some markets in Awka, Nigeria. J Biosci Bioeng 3:90–94. https://doi.org/10.13189/bb.2015.030503
Shehu K, Muhammad S (2012) Fungi associated with storage rots of onion bulbs in Sokoto, Nigeria. Int J Mod 1:1–3. https://doi.org/10.5923/j.ijmb.20110101.01
Susca A, Proctor RH, Morelli M, Haidukowski M, Gallo A, Logrieco AF, Moretti A (2016) Variation in fumonisin and ochratoxin production associated with differences in biosynthetic gene content in Aspergillus niger and A. welwitschiae isolates from multiple crop and geographic origins. Front Microbiol 7:1–16. https://doi.org/10.3389/fmicb.2016.01412
Susca A, Proctor RH, Mulè G, Stea G, Ritieni A, Logrieco A, Moretti A (2010) Correlation of mycotoxin fumonisin B2 production and presence of the fumonisin biosynthetic gene fum8 in Aspergillus niger from grape. J Agric Food Chem 58:9266–9272. https://doi.org/10.1021/jf101591x
Susca A, Proctor RH, Butchko RAE, Haidukowski M, Stea G, Logrieco A, Moretti A (2014) Variation in the fumonisin biosynthetic gene cluster in fumonisin-producing and nonproducing black aspergilli. Fungal Genet Biol 73:39–52. https://doi.org/10.1016/j.fgb.2014.09.009
Olarte RA, Menke J, Zhang Y, Sullivan S, Slot JC, Huang Y, Badalamenti JP, Quandt AC, Sparafora JW, Bushley KE (2019) Chromosome rearrangements shape the diversification of secondary metabolism in the cyclosporin producing fungus Tolypocladium inflatum. BMC Genomics 20:120. https://doi.org/10.1186/s12864-018-5399-x
Turner G (2010) Genomics and secondary metabolism in Aspergillus. In: Machida M, Gomi K (eds) Aspergillus: molecular biology and genomics. Caister Academic Press, Norfolk, pp 139–155
Ferracin LM, Fier CB, Vieira MLC, Monteiro-Vitorello CB, Varani A de M, Rossi MM, Muller-Santos M, Taniwaki MH, Iamanaka BT, MHP F (2012) Strain-specific polyketide synthase genes of Aspergillus niger. Int J Food Microbiol 155:137–145. https://doi.org/10.1016/j.ijfoodmicro.2012.01.020
Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, de Vries RP, Albang R, Albermann K, Andersen MR, Bendtsen JD, Benen JA, van den Berg M, Breestraat S, Caddick MX, Contreras R, Cornell M, Coutinho PM, Danchin EG, Debets AJ, Dekker P, van Dijck PW, van Dijk A, Dijkhuizen L, Driessen AJ, d'Enfert C, Geysens S, Goosen C, Groot GS, de Groot PW, Guillemette T, Henrissat B, Herweijer M, van den Hombergh JP, van den Hondel CA, van der Heijden RT, van der Kaaij RM, Klis FM, Kools HJ, Kubicek CP, van Kuyk PA, Lauber J, Lu X, van der Maarel MJ, Meulenberg R, Menke H, Mortimer MA, Nielsen J, Oliver SG, Olsthoorn M, Pal K, van Peij NN, Ram AF, Rinas U, Roubos JA, Sagt CM, Schmoll M, Sun J, Ussery D, Varga J, Vervecken W, van de Vondervoort PJ, Wedler H, Wosten HA, Zeng AP, van Ooyen AJ, Visser J, Stam H (2007) Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol 25:221–231. https://doi.org/10.1038/nbt1282
Frisvad JC, Larsen TO, Thrane U, Meijer M, Varga J, Samson RA, Nielsen KF (2011) Fumonisin and ochratoxin production in industrial Aspergillus niger strains. PLoS One 6:2–7. https://doi.org/10.1371/journal.pone.0023496
Castellá G, Cabanes FJ (2011) Development of a real time PCR system for detection of ochratoxin A-producing strains of the Aspergillus niger aggregate. Food Control 22:1367–1372. https://doi.org/10.1016/j.foodcont.2011.02.014
Yamada O, Machida M, Hosoyama A, Goto M, Takahashi T, Futagami T, Yamagata Y, Takeuchi M, Kobayashi T, Koike H, Abe K, Asai K, Arita M, Fujita N, Fukuda K, Higa K, Horikawa H, Ishikawa T, Jinno K, Kato Y, Kirimura K, Mizutani O, Nakasone K, Sano M, Shiraishi Y, Tsukahara M, Gomi K (2016) Genome sequence of Aspergillus luchuensis NBRC 4314. DNA Res 23(6):507–515. https://doi.org/10.1093/dnares/dsw032
Funding
This study was financed by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) (Project 2013/05414-8), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) (F.M. Pelisson, grant Code 001), and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPQ) (M. H. P. Fungaro, grant #310970/2015-6).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no conflict of interest.
Additional information
Responsible Editor: Rosana Puccia.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Highlights
• Aspergillus welwitschiae is very frequently found in onions marketed in Brazil.
• An essential gene involved in fumonisin biosynthesis (fum8) is present in 36% of Aspergillus welwitschiae strains.
Rights and permissions
About this article
Cite this article
Massi, F.P., Iamanaka, B.T., Barbosa, R.L. et al. Molecular analysis of Aspergillus section Nigri isolated from onion samples reveals the prevalence of A. welwitschiae. Braz J Microbiol 52, 387–392 (2021). https://doi.org/10.1007/s42770-020-00390-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-020-00390-2