Abstract
Plant growth-promoting bacteria such as Streptomyces are an attractive alternative for increasing the sustainability of agricultural systems. In this study, Streptomyces isolates obtained from rhizosphere soil of plants in the family Fabaceae were characterized for their plant growth-promoting traits, including the production of siderophores, 1-aminocyclopropane-1-carboxylate (ACC) deaminase, indole-3-acetic acid (IAA), and phenazines. Soybean seeds were bacterized with selected isolates to test growth promotion. All isolates produced IAA, and the isolate CLV45 was the most efficient, reaching 398.53 mg of IAA per gram of cells. CLV41, CLV45, and CLV46 showed high activity for ACC deaminase whereas CLV42, CLV44, and CLV46 were efficient in siderophore production. Pyocyanin was detected in all isolates; CLV41, CLV43, and CLV45 produced phenazine-carboxylic acid as well. Selected for IAA and ACC deaminase production combined with production of siderophores and phenazines, CLV42, CLV44, and CLV45 were tested for their growth promotion potential. Seed bacterization with CLV45 resulted in plants with increased shoot growth (36.63%) and dry mass (17.97%) compared to control plants. Results suggest that moderate or high levels of auxin and ACC deaminase production by the isolate CLV45 positively affected the growth of soybean plants, making it a strong candidate for further studies on biofertilizer formulation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bacteria that colonize roots and promote plant growth and health are termed plant growth-promoting rhizobacteria (PGPR). Generally, PGPR benefit plant growth either by assisting in nutrient availability (nitrogen, phosphorus, and iron) or by producing phytohormones such as indole acetic acid (IAA) [1]. PGPR can also indirectly help plants to withstand stress (biotic or abiotic) by reducing ethylene levels, through production of enzyme 1-aminocyclopropane-1-carboxylic acid deaminase (ACC deaminase) [2, 3]; by locally producing antagonists to soil-borne pathogens; or by inducing systemic resistance (ISR) through plant recognition of microorganism-associated molecular patterns (MAMPs) [4, 5]. Indeed, PGPR may use one or a combination of these mechanisms and act as biofertilizers, phytostimulators, and biocontrol agents in the rhizosphere where they are established [6, 7].
In current agricultural practice, the input of fertilizers has increased to maintain the growth of important crops. However, the excessive use of agrochemicals on crop fields has been reported to increase nitrate, nitrite, ammonium, and phosphate contents, as well as other reactive chemical species in groundwater and surface water bodies, which may cause serious environmental and health hazards [7]. The use of PGPR as microbial inoculants is one alternative for sustainable agriculture [8].
Among PGPR, some strains of the Gram-positive filamentous actinobacteria Streptomyces have been reported as having a vast potential to stimulate plant growth and induce defense responses in plants colonized by these microorganisms [9]. There is also great interest in these bacteria due to their potential to produce antimicrobial metabolites such as phenazines [4, 10]. Strains of Streptomyces species originating from an Araucaria forest were characterized as PGPR, based on their capacity for IAA production [11], phosphate solubilization, antibiosis, and production of siderophores [12]. Some studies have shown a positive effect of Streptomyces spp. on the growth of different crop species, such as wheat [13] and tomato [3, 12]. In eucalyptus plants, inoculation of roots with Streptomyces sp. PM9 resulted in a significant increase of secondary roots and modulation of secondary metabolism [14]. Although some reports on actinobacteria, namely Streptomyces, have proven their positive effects on plant growth, their potential as PGPR is still underexploited [15].
Soybean (Glycine max (L.) Merr.), a dicotyledonous plant in the family Fabaceae, is one of the most important crops for seed protein and oil content [16]. As with many other crops, the management of this species requires the use of chemical fertilizers. In the search for a more sustainable agriculture, the combination of cultivated plants with PGPR can yield economic and environmental benefits. However, root colonization and consequent plant growth might involve mechanisms of host recognition and control of bacterial properties by plant exudates [17]. Thus, we hypothesized that Streptomyces species isolated from the rhizosphere of members of the Fabaceae family could associate with soybean plants and positively affect the growth of this crop.
This study biochemically characterized 11 isolates of Streptomyces obtained from soil and the roots of Fabaceae species regarding their capacity to produce siderophores, IAA, phenazines, and the enzyme ACC deaminase. Based on these PGPR traits, the effects of three strains of Streptomyces on the growth of soybean plants were analyzed.
Material and methods
Microorganisms and culture conditions
Fragments of roots and rhizospheric soil from different plants in the family Fabaceae were collected in several localities in Rio Grande do Sul, Brazil (Table 1). Isolation of rhizobacteria followed procedures for Streptomyces selection. Briefly, rhizobacteria were obtained from rhizospheric soil by incubating the samples in HCN liquid medium [18] at 100 rpm and 42 °C for 30 min, followed by culturing on ISP2 medium [19] supplemented with the antibiotics cycloheximide (100 μg mL−1) and nalidixic acid (50 μg mL−1) and the antifungal nystatin (100 μg mL−1). Eleven isolates were initially identified as Streptomyces species based on their morphology [20] and stored at − 80 °C in a 20% glycerol solution. Preparation of the isolates as inocula for the assays (characterization as PGPR and molecular identification) was carried out in ISP2 liquid medium under agitation at 100 rpm for 5 days at 26 ± 2 °C (log phase). Each suspension was centrifuged (2500g, 15 min, room temperature), resuspended in sterile distilled water, and adjusted to a final concentration of 107–108 CFU mL−1.
Biochemical characterization of the isolates
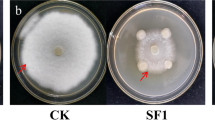
All isolates were evaluated for PGPR characteristics, including the production of siderophores, ACC deaminase, IAA, and phenazines. To assess the siderophore production ability, Chrome Azurol S (CAS) agar plates were prepared according to the cup-plate method (5-mm wells) [21]. Each isolate was grown in ISP2 medium (10 mL) for 5 days at 26 ± 2 °C and inoculated onto three plates of CAS-LB agar, each with three wells (100 μL per well), totaling nine samples per isolate. The plates were incubated at 28 ± 2 °C for 7 days. Distilled water and pyrocatechol (1 M) were used as negative and positive controls, respectively. When siderophores were released by the bacteria, the color of the medium changes from blue to yellowish orange (Fig. 1). The diameter of the halo zone (cm) after incubation was measured. Data were expressed as the mean of the diameter of halo boundary ± standard error (SE).
The presence of ACC deaminase (E.C. 4.1.99.4) produced by the Streptomyces isolates was determined following Cattelan et al. [22], with modifications. The isolates were previously cultured in 10 mL ISP2 liquid medium for 5 days at 26 ± 2 °C, centrifuged (20 min, 2500g), and washed twice with Dworkin and Foster (DF) minimal salt medium [23] without glucose or nitrogen (N) salts. The pellet was resuspended in 10 mL of liquid DF salt medium without N and agitated for 2 days at 100 rpm and 26 ± 2 °C. The ACC deaminase assay comprised three treatments using media with different N sources. In a 24-well plate, 500 μL of semi-solid DF salt medium with ACC (6 mg mL−1) as the only N source was dispensed aseptically into each well (DF+ACC). In another multi-well plate, the same medium was prepared without any sources of N (DF-N). For positive control, an identical plate was prepared with DF salt medium containing N (DF complete, DF+N). Aliquots (5 μL) of the isolates were cultured in duplicate per treatment, with sterile distilled water used as the control. Plates were incubated at 28 ± 2 °C for 5 days. The qualitative assessment was based on the growth of each isolate, which was considered positive for ACC deaminase production if the isolate grew on the medium with ACC and showed no growth on DF-N.
Auxin production was analyzed by Salkowski’s method [24], with modifications. The isolates were previously grown in 10 mL liquid ISP2 medium for 5 days at 26 ± 2 °C under agitation at 100 rpm. Bacterial suspensions were centrifuged for 15 min at 2500g at room temperature. The supernatant was combined with Salkowski’s reagent (1:1, v/v) and incubated for 30 min at room temperature in the dark. Absorbances were read in a spectrophotometer at 530 nm from five replicates per isolate, and IAA levels were estimated in relation to the standard calibration curve of the hormone. The mass of the bacterial pellet was measured to calculate the IAA concentration per gram of cells.
Phenazines were identified and quantified using Streptomyces isolates grown in 10 mL ISP2 liquid medium at 100 rpm and 26 ± 2 °C for 7 days (decline phase). Extraction and characterization procedures followed that of Kadam et al. [25] and Cezairliyan et al. [26], with modifications. For extraction of phenazines, 20 mL of each isolate suspension was centrifuged, and the supernatant was divided equally into two flasks. Pellets with bacterial cells were weighed. Purification of pyocyanin (PYO) was performed by adding chloroform (1:1, v/v) to the supernatant. The solution was gently mixed for 30 min and then acidified to pH 2 using concentrated HCl. The organic phase was used for analysis at 365 nm. Phenazine-1-carboxylic acid (PCA) and 1-hydroxyl-phenazine (OH-Phe) were extracted by acidifying the supernatant with 6 M HCl (1:1, v/v), followed by addition of ethyl acetate (1:1, v/v). The mixture was gently agitated and allowed to stand for 30 min for phase separation. The organic phase was evaporated under air stream, and the residue was resuspended with 200 μL of 1 M NaOH. The same volume of methanol was added to the solution, and the analysis was performed at 365 nm. The analytical HPLC system comprised a Sykam Research HPLC S 600 chromatography system with a UV/VIS detector Mod. 3345 DAD. Phenazine separation was done in a MetaSil ODS column (5 μm, 250 × 4.6 mm) with a C18 guard column. The temperature of the column oven was set at 40 °C. Chromatographic data were processed by Clarity Chromatography Software. Mobile phase A consisted of water, and mobile phase B consisted of acetonitrile. Both eluents were acidified using 2.5% formic acid. The linear gradient consisted of 0–15% of eluent B for 2 min, 15–83% of B for 12 min, 83–0% of B for 2 min, and 0% of B for 4 min under a flow rate of 1 mL min−1, as modified from Kern and Newman [27]. Quantification was based on calibration curves of the standards obtained from the manufacturers: 1-Hydroxyphenazine was obtained from Tokyo Chemical Industry Co. (Japan), pyocyanin from Sigma Chemical Co. (USA), and phenazine-1-carboxylic acid from iChemical (China). The concentration was expressed as micrograms per gram of phenazine cells. Two experimental replicates were used for each standard and bacterial sample.
Selection of Streptomyces isolates for plant growth experiment
Three isolates of Streptomyces were selected based on the hypothesis that promotion of plant growth is a consequence of a combination of metabolic characteristics rather than one PGPR trait. The criterion for selection was efficient production of IAA, combined with differential production of ACC deaminase by the isolates, including the absence of this enzyme. A Streptomyces isolate with highly positive production of siderophores was also selected, regardless of other parameters. Production of phenazines was also considered in the choice of isolates.
Molecular identification of the selected isolates and phylogenetic analysis
Isolates selected for their biochemical PGPR characteristics were taxonomically identified using 16S rRNA gene sequencing. Pure cultures in 10 mL liquid ISP2 medium were prepared and incubated at 26 °C for 3 days (exponential phase). An aliquot of 100 μL of each bacterial suspension was plated on semi-solid ISP2 for 4 days. Bacterial DNA was extracted using the Wizard® Genomic DNA Purification Kit (Promega Biotecnologia do Brasil, Ltda.). PCR amplification of the 16S rRNA gene was performed using the universal primers 9F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1542R (5′-AGAAAGGAGGTGATCCAGCC-3′), and PCR-amplified fragments were sequenced by Macrogen Inc. (Republic of Korea). The sequences obtained and another 20 Streptomyces 16S rRNA sequences from different species randomly retrieved from the NCBI database (National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/) were used to perform the phylogenetic analysis. Sequences were aligned using CLUSTAL W. To ensure the stability and reliability of the phylogenetic relationships among the strains used in this study, a phylogenetic tree was constructed through the maximum likelihood (ML) method and Tamura-Nei model, using MEGA X. The topology of the phylogenetic trees was evaluated by bootstrap resampling (1000 replications).
Promotion of plant growth by Streptomyces spp.
Soybean seeds (TMG-7262 RR INOX) with no fungicide treatment were provided by Ballagro AgroTecnologia Ltda., Brazil. For use in the experiments, seeds were surface-disinfested with sodium hypochlorite solution (2%, v/v) for 2 min, rinsed three times with sterile distilled water, and treated with the isolates. The procedure was carried out as follows: three selected isolates (CLV42, CLV44, and CLV45) were grown in 10 mL ISP2 liquid medium, under 100 rpm, at 26 °C. After 5 days of culture, the bacterial suspension was adjusted to the final concentration of 107–108 CFU mL−1. For the bacterization, ten seeds were immersed in 5 mL of bacterial suspension per treatment (CLV42, CLV44, and CLV45) for 5 min and partially dried for 1 h in uncovered Petri dishes in a laminar flow hood. For the control, seeds were kept in sterile distilled water and dried under the same conditions. Seeds were sown in non-sterile commercial organic soil (13% clay, 7.7% organic matter and ground calcareous rock; pH 6.6 (measured in water 1:5 (w/v)); 260 μS cm−1) in polypropylene bags (15 × 25 cm) and transferred to a greenhouse (photoperiod 18 h and 23–30 °C). After thinning, the experiment was carried out with 30 plants per treatment and control group. The plants were irrigated with tap water when necessary. Nutrient solution (10 mL of macronutrient salt solution, containing (per liter) 0.41 g NH4NO3, 0.47 g KNO3, 110 g CaCl2·2H2O, 0.09 g MgSO4·7H2O, and 0.04 g KH2PO4) was supplied to the plants every 15 days. Soybean plant growth was evaluated based on length (cm) as well as fresh and dry mass (g) of the shoots and roots at 15, 30, and 45 days after sowing. Dry mass was determined by oven drying roots and shoots at 60 °C to constant mass.
Statistical analysis
Experiments were performed in a fully randomized design, tested for variance homogeneity by Levene’s test (α ≤ 0.05). Data from experiments of biochemical characterization of Streptomyces spp. were analyzed by one-way ANOVA, and means were compared by Duncan’s multiple range test at a significance level of α ≤ 0.05. The results obtained in the growth promotion assay were analyzed by Student’s t test (α ≤ 0.05). All statistical analyses were performed using the software SPSS Statistics v. 22. Data from experiments were expressed as mean ± SE.
Results
Biochemical characterization
The 11 bacterial isolates obtained from the rhizosphere of plants of the family Fabaceae and morphologically identified as Streptomyces were biochemically characterized to determine their potential as PGPR. All isolates were able to produce siderophores under iron-limiting conditions (Table 2). Isolate CLV42 showed the highest siderophore production, followed by CLV46, CLV44, and CLV41 (Fig. 1, Table 2). On the other hand, the presence of ACC deaminase was not ubiquitous in the isolates. Five of the 11 Streptomyces sp. isolates showed growth on the medium supplemented with ACC as the only source of N, indicating activity of the enzyme ACC deaminase (Table 2). Different intensities of enzyme activity were observed among these Streptomyces isolates, and CLV41, CLV45, and CLV46 showed the most efficient growth during the culture period of 5 days (Table 2).
All isolates were able to produce IAA (Table 2), although production differed among the isolates (6.48–398.53 μg g−1 of cells). Isolate CLV45 showed the highest IAA content, followed by isolates CLV43, CLV44, and CLV40, whereas isolate CLV41 produced the smallest amount of this hormone (Table 2).
Production of phenazines PYO, PCA, and OH-Phe was evaluated in the supernatants of the cultures (Fig. 2a). All isolates were able to produce PYO. The most productive isolate was CLV26 (450.51 μg g−1 cell), followed by isolates CLV21 (305.28 μg g−1 cell) and CLV43 (225.61 μg g−1 cell) (Fig. 2b). On the other hand, only some isolates produced PCA (Fig. 2b). The highest concentration of PCA was found in the supernatant of the CLV43 culture, although this level was 23.6 times lower than the production of PYO by the same isolate. OH-Phe was not detected in any of the isolates tested.
Phenazine production. a Chromatogram of phenazines: A. PYO (pyocyanin); B. PCA (phenazine-1-carboxylic acid); C. OH-phenazine; D. phenazine (basic molecule). b Quantification of phenazines, PYO and PCA, in the supernatant of Streptomyces spp. isolates cultures. Samples were evaluated in duplicates by HPLC
Selection of isolates for molecular analysis and growth promotion assay
The Streptomyces isolates were chosen for their performance in the metabolic evaluation of their PGPR traits, such as IAA and ACC deaminase production. CLV42, CLV44, and CLV45 were selected for low, moderate, and high production of IAA, respectively. Likewise, ACC deaminase was undetectable in CLV42, and CLV44 and CLV45 showed weak and strong colony growth in the presence of ACC, respectively. On the other hand, CLV42 produced more siderophores than the other isolates (Table 2). All three isolates produced PYO, and CLV45 produced both PYO and PCA. Hence, these three isolates were selected for the experiment to evaluate the ability of Streptomyces spp. to promote the growth of soybean plants.
The 16S rRNA partial gene sequences of the three selected isolates were compared with 16S rRNA sequences of Streptomyces species in the NCBI GenBank database. Analysis showed that the three isolates belonged to the genus Streptomyces (Fig. 3), and their sequences were deposited in GenBank under accession numbers CLV42–KY704165, CLV44–KY704108, and CLV45–KY704164. The phylogeny showed that the isolates grouped in three distinct clades. It also indicated that CLV42 and CLV45 are more closely related to each other than to CLV44. The position of isolate CLV42 indicated a high similarity to Streptomyces muensis. The CLV45 placement showed that it may be related to Streptomyces globisporus and Streptomyces pulveraceus, which are all equally linked to Streptomyces gelaticus. CLV44 was placed within a more distant clade, in which it appears related to both Streptomyces atratus and Streptomyces sanglieri (Fig. 3).
Taxonomic identification of bacterial isolates CLV42–KY704165, CLV44–KY704108, and CLV45–KY704164. Phylogenetic analyses were performed with reference sequences obtained from the NCBI GenBank database. Phylogenetic trees were constructed using the maximum likelihood method and Tamura-Nei model based on 16S rRNA partial gene sequences. Bootstrap percentages based on 1000 replications are shown at branch points
Promotion of plant growth by Streptomyces spp.
Bacterization of soybean seeds with Streptomyces isolates CLV44 and CLV45 influenced plant growth differently during the culture (Fig. 4). At 15 days, soybean plants treated with CLV44 and CLV45 had shorter roots and shoots compared to the control plants (Fig. 4a, b). The fresh and dry mass of roots also differed significantly from the control (Fig. 4c, e). However, at 30 days from the beginning of the experiment, soybean plants bacterized with Streptomyces sp. CLV44 showed significantly longer shoots and higher dry mass compared to the control group (Fig. 4b, f). Despite this early increased growth, at 45 days, the plants treated with isolate CLV44 showed equal growth to the control.
Evaluation of Streptomyces sp. CLV42, CLV44, and CLV45 isolates on the promotion of soybean plant growth. a Root length (cm). b Shoot length (cm). c Fresh root mass (mg). d Fresh shoot mass (mg). e Dry root mass (mg). f Dry shoot mass (mg). Different letters within each time of analysis mean significant difference by t test (α ≤ 0.05)
On the other hand, despite the negative or neutral effect of isolate CLV45 on growth at early time points (15 and 30 days), a significant positive effect was observed on soybean plants at 45 days, with a 36.63% increase in shoot growth and 17.97% in shoot dry weight in relation to the control (Fig. 4b, f).
Treatment of plants with Streptomyces sp. CLV42 resulted in no statistical difference in growth during the culture period compared to the control group and was therefore considered to have a neutral effect on soybean seeds/plants.
Discussion
In recent decades, several studies have demonstrated the potential of plant growth-promoting rhizobacteria (PGPR) for improving crop performance and yield and for controlling pathogen attack [1, 6, 8]. Among rhizobacteria, Streptomyces have been reported as PGPR, and despite strong interest in the medical field because of their high production of secondary metabolites, few studies have examined this group as biofertilizers or biocontrol agents [12, 14, 28,29,30,31,32]. The beneficial effects of these rhizobacteria have been attributed to their ability to produce several compounds including phytohormones, siderophores, antibiotics, and lytic enzymes, as well as to fix atmospheric nitrogen, solubilize phosphate, and induce systemic resistance.
In the present study, all 11 isolates of Streptomyces obtained from the rhizosphere of species of Fabaceae showed at least two characteristics of PGPR. All isolates produced siderophores and IAA. Specifically, isolate CLV42 was the most efficient in producing siderophores, and CLV45 produced the most IAA (398.5 μg g−1 of cells), resulting in 3.65 times more hormone than the second-best producer, CLV44. Production of siderophores and IAA is a common feature of plant growth-promoting rhizobacteria. Production of bacterial siderophores stimulates plant growth by increasing iron availability in the rhizosphere and has been previously reported in Streptomyces species [13, 33, 34]. IAA is the main auxinic hormone produced by rhizobacteria [2], and its production is associated with remodeling and development of the roots, increasing nutrient absorption by the plant, and thus promoting growth and stress tolerance [35]. Plant growth responses related to Streptomyces spp. IAA producers have been reported in important crops such as rice [36], tomato [12], wheat [13], and eucalyptus [14, 37]. Notably, most of the isolates analyzed in this study showed IAA production lower than 50 μg g−1 of cells, a concentration that has been used by some companies to formulate their fertilizers (PuraKelp; Omnia Nutriologia, Brazil). Based on this criterion, the isolates with low production of IAA were considered inefficient producers for commercial application.
The presence of ACC deaminase was more limited in the isolates of Streptomyces compared to the characteristics discussed above, i.e., was recorded in only 5 of the 11 isolates. ACC deaminase is an enzyme that can metabolize ACC, the precursor of ethylene, thereby reducing the excess of this hormone. Once ethylene levels are reduced in the rhizosphere by PGPR, increased root elongation and plant growth are expected [2, 38]. ACC deaminase-producing strains of Streptomyces sp. showed an effect on alleviation of salt stress in tomato and rice plants, respectively [3, 39]. Our results showed that Streptomyces sp. CLV45 was an efficient IAA producer and grew strongly in the presence of ACC as the only source of nitrogen, traits that make it a candidate for further studies on the growth of soybean plants.
Isolates of Streptomyces spp. were also capable of producing phenazines. Although PCA was found only in the supernatant of CLV41, CLV43, and CLV45 cultures, PYO was detected in all isolates and in very high concentrations in some (CLV21 and CLV26). Phenazines are a diverse class of heterocyclic secondary metabolites that have been studied for many years due to their antibiotic properties and role in virulence [40]. Most plants greatly benefit from phenazine production by bacteria since these molecules can inhibit fungal and bacterial pathogens with PYO and PCA, involved in induced systemic resistance [41, 42]. Moreover, bacterial production of PYO confers advantages such as enhancing bacterial adhesion, microcolony formation, and increasing biomass, as reported for Pseudomonas aeruginosa [43]. With these biological properties, phenazines from Streptomyces might exert an indirect effect on plant growth, favoring the production of biofilms and promoting bacterial colonization and persistence on plant roots. Root colonization by PGPR improved by biofilm formation has been described in barley [44].
Considering the growth promotion traits shown by the different Streptomyces species, three isolates were selected to test their effect on the growth of soybean plants. At day 15, bacterization with isolates CLV44 and CLV45 caused a decrease in the growth parameters analyzed compared to control (non-bacterized) plants. This initial performance of Streptomyces sp. CLV44 and CLV45 on soybean seedlings may be related to the high production of IAA by these isolates (109.04 and 398.53 μg g−1 cells, respectively), which may have negatively affected the development of tap roots during germination and early seedling development. It is well known that high concentrations of IAA inhibit the elongation and growth of the tap root. Moreover, the IAA produced by the Streptomyces isolates could also be taken up by the root during initial development, most likely stimulating the activity of the enzyme ACC synthase, which converts S-adenosylmethionine to ACC. Consequently, the increase in the substrate for ACC oxidase elevates the endogenous concentrations of ethylene [2] and ultimately inhibits root growth. Furthermore, the contact of the rhizobacteria with the developing roots can cause a delay in the initial plant growth due to the deviation in the cellular metabolism to recognize and establish the rhizobacteria-plant interaction, a trade-off often reported in this type of interaction [11, 14, 45].
Although some negative effect of bacterization on plant growth was observed in the first 15 days of the experiment, this changed over the culture period, and at 45 days, promotion of plant growth mediated by CLV45 was evident. Compared to the control plants, shoot growth and shoot dry weight increased 36.63% and 17.97%, respectively. This result could be attributed to an indirect role of the IAA secreted by a rhizobacterium and its interaction with ethylene in promoting shoot growth. The model proposed by Glick [46] states that a complex exchange of signals between IAA and ethylene in plant growth promotion by PGPR can occur. Thus, the coordinated production of IAA and ACC deaminase by the CLV45 isolate may have been responsible for stimulating the growth of the soybean plants. We also suggest that the capability of producing PYO might have contributed to root colonization by promoting bacteria adhesion to plant roots. The successful interaction between Streptomyces CLV45 and soybeans may have been potentiated by the natural interaction between this isolate and Phaseolus vulgaris. Soybean and Phaseolus are related, both belonging to subfamily Faboideae.
Few studies have described Streptomyces species as PGPR, and even fewer have evaluated Streptomyces from soil samples in South America [14, 28]. Surveys that isolated and identified actinobacteria (mainly Streptomyces species) from Brazilian soils have found a rich, though largely still unknown, group possibly with high biotechnological potential including use as PGPR [47, 48]. To the best of our knowledge, none of the species that clustered with Streptomyces CLV42, CLV44, and CL45 in our phylogenetic analysis has been reported as PGPR to date.
The present data contribute significantly to unveil this diversity of Streptomyces species and their abilities as PGPR for soybean, an economically important crop in Brazil and many other countries.
Conclusion
The isolates of Streptomyces species collected from the rhizosphere of members of the Fabaceae family showed different plant growth-promoting characteristics that can be used to improve plant growth. The greenhouse growth promotion trial showed that the Streptomyces isolate CLV45 promoted the growth of soybean plants and indicated that it is a strong candidate for biofertilizer formulation. None of the isolates evaluated in greenhouse conditions caused a growth deficit in soybean plants. A consortium combining different isolates of Streptomyces may have good potential as a plant biofertilizer since the combined PGPR characteristics of each isolate may benefit crops and the environment by reducing the use of fertilizers and increasing productivity. Although research with streptomycetes as PGPR and their use as bioinoculants has increased in recent decades, it is still necessary to improve knowledge on the diversity of metabolites produced by different strains of Streptomyces in order to exploit the maximum potential of this group for use in high-impact crops such as soybeans.
References
Gopalakrishnan S, Srinivas V, Alekhya G, Prakash B et al (2015) The extent of grain yield and plant growth enhancement by plant growth-promoting broad-spectrum Streptomyces sp. in chickpea. SpringerPlus 4:31
Glick BR (2014) Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol Res 169:30–39
Palaniyandi SA, Damodharan K, Yang SH, Suh JW (2014) Streptomyces sp. strain PGPA39 alleviates salt stress and promotes growth of “Micro Tom” tomato plants. J Appl Microbiol 117:766–773
Luo Q, Hu H, Peng H, Zhang X, Wang W (2015) Isolation and structural identification of two bioactive phenazines from Streptomyces griseoluteus P510. Chinese J Chem Eng 23:699–703
Mhlongo MI, Piater LA, Madala NE, Labuschagne N, Dubery IA (2018) The chemistry of plant–microbe interactions in the rhizosphere and the potential for metabolomics to reveal signaling related to defense priming and induced systemic resistance. Front Plant Sci 9:112. https://doi.org/10.3389/fpls.2018.00112
Bhattacharyya PN, Jha DK (2012) Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture. World J Microbiol Biotechnol 28:1327–1350
Gupta G, Parihar SS, Ahirwar NK, Snehi SK, Singh V (2015) Plant growth promoting rhizobacteria (PGPR): current and future prospects for development of sustainable agriculture. J Microb Biochem Technol 7:96–102
Etesami H, Maheshwari DK (2018) Use of plant growth promoting rhizobacteria (PGPRs) with multiple plant growth promoting traits in stress agriculture: action mechanisms and future prospects. Ecotoxicol Environm Saf 156:225–246
Bishnoi U (2015) PGPR interaction: an ecofriendly approach promoting the sustainable agriculture system. In: Advances in botanical research, vol 75. Academic Press, pp 81–113
Wang Y, Luo Q, Zhang X, Wang W (2011) Isolation and purification of a modified phenazine, griseoluteic acid, produced by Streptomyces griseoluteus P510. Res Microbiol 162:311–319
Dalmas FR, Pereira TCB, Bogo MR, Astarita LV (2011) Autochthonous Streptomyces regulate the metabolism of seedlings of Araucaria angustifolia (Coniferales) during root colonisation. Aust J Bot 59:118–125
Dias MP, Bastos MS, Xavier VB, Cassel E, Astarita LV, Santarém ER (2017) Plant growth and resistance promoted by Streptomyces spp. in tomato. Plant Physiol Biochem 118:479–493
Sadeghi A, Karimi E, Dahaji PA, Javid MG, Dalvand Y, Askari H (2012) Plant growth promoting activity of an auxin and siderophore producing isolate of Streptomyces under saline soil conditions. World J Microbiol Biotechnol 28:1503–1509
Salla TD, Ramos da Silva T, Astarita LV, Santarém ER (2014) Streptomyces rhizobacteria modulate the secondary metabolism of Eucalyptus plants. Plant Physiol Biochem 85:14–20
Gong Y, Bai JL, Yang HT, Zhang WD, Xiong YW, Ding P, Qin S (2018) Phylogenetic diversity and investigation of plant growth-promoting traits of actinobacteria in coastal salt marsh plant rhizospheres from Jiangsu, China. Syst Appl Microbiol 41:516–527. https://doi.org/10.1016/j.syapm.2018.06.003
Mingma R, Pathom-aree W, Trakulnaleamsai S, Thamchaipenet A, Duangmal K (2014) Isolation of rhizospheric and roots endophytic actinomycetes from Leguminosae plant and their activities to inhibit soybean pathogen Xanthomonas campestris pv. glycine. World J Microbiol Biotechnol 30:271–280
Drogue B, Doré H, Borland S, Wisniewski-Dyé F, Prigent-Combaret C (2012) Which specificity in cooperation between phytostimulating rhizobacteria and plants? Res Microbiol 163(8):500–510
Nonomura H, Hayakawa M (1988) New methods for the selective isolation of soil actinomycets. In: Okami Y, Beppu T, Ogawara H (eds) Biology of actinomycetes 88: proceedings of seventh international symposium on biology of actinomycetes. Japan Scientific Societies Press, Tokyo, pp 288–293
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16:313–340
Dhanasekaran D, Jiang Y (2016) Actinobacteria—basics and biotechnological applications. IntechOpen, London, p 387
Dingle J, Reid WW, Solomons GL (1953) The enzymic degradation of pectin and other polysaccharides. II-Application of the ‘cup-plate’ assay to the estimation of enzymes. J Sci Food Agric 4:149–155
Cattelan AJ, Hartel PG, Fuhrmann JJ (1999) Screening for plant growth-promoting rhizobacteria to promote early soybean growth. Soil Sci Soc Am J 63:1670–1680
Dworkin M, Foster JW (1958) Experiments with some microorganisms which utilize ethane and hydrogen. J Bacteriol 75:592–603
Salkowski E (1885) Ueber das verhalten der skatolcarbonsäure im organismus. Zeitschr Physiol Chem 9:23–33
Kadam MS, Patil SG, Dane PR, Pawar MK, Chincholkar SB (2013) Methods for purification and characterization of microbial phenazines. In: Microbial Phenazines. Springer, Berlin, Heidelberg, pp 101–140
Cezairliyan B, Vinayavekhin N, Grenfell-Lee D, Yuen GJ, Saghatelian A, Ausubel FM (2013) Identification of Pseudomonas aeruginosa phenazines that kill Caenorhabditis elegans. PLoS Pathog 9:e1003101
Kern SE, Newman DK (2014) Measurement of phenazines in bacterial cultures. In: Filloux A, Ramos J-L (eds) Pseudomonas methods and protocols, vol 1149. Springer, New York, pp 303–310
Salla TD, Astarita LV, Santarém ER (2016) Defense responses in plants of Eucalyptus elicited by Streptomyces and challenged with Botrytis cinerea. Planta 243:1055–1070
Tamreihao K, Ningthoujam DS, Nimaichand S, Singh ES, Reena P, Singh SH, Nongthomba U (2016) Biocontrol and plant growth promoting activities of a Streptomyces corchorusii strain UCR3-16 and preparation of powder formulation for application as biofertilizer agents for rice plant. Microbiol Res 192:260–270
Rey T, Dumas B (2017) Plenty is no plague: Streptomyces symbiosis with crops. Trends Plant Sci 22:30–37
Patel JK, Madaan S, Archana G (2018) Antibiotic producing endophytic Streptomyces spp. colonize above-ground plant parts and promote shoot growth in multiple healthy and pathogen-challenged cereal crops. Microbiol Res 215:36–45. https://doi.org/10.1016/j.micres.2018.06.003
Singh RP, Manchanda G, Maurya IK, Maheshwari NK, Tiwari PK, Rai AK (2019) Streptomyces from rotten wheat straw endowed the high plant growth potential traits and agro-active compounds. Biocatal Agricult Biotechn 17:507–513. https://doi.org/10.1016/j.bcab.2019.01.014
Rajkumar M, Ae N, Prasad MNV, Freitas H (2010) Potential of siderophore-producing bacteria for improving heavy metal phytoextraction. Trends Biotechnol 28:142–149
Palaniyandi SA, Yang SH, Zhang L, Suh JW (2013) Effects of actinobacteria on plant disease suppression and growth promotion. Appl Microbiol Biotechnol 97:9621–9636
Egamberdieva D, Wirth SJ, Alqarawi AA, Abd Allah EF, Hashem A (2017) Phytohormones and beneficial microbes: essential components for plants to balance stress and fitness. Front Microbiol 8. https://doi.org/10.3389/fmicb.2017.02104
Gopalakrishnan S, Vadlamudi S, Bandikinda P, Sathya A, Vijayabharathi R, Rupela O, Kudapa H, Katta K, Varshney RK (2014) Evaluation of Streptomyces strains isolated from herbal vermicompost for their plant growth-promotion traits in rice. Microbiol Res 169:40–48
Mafia RG, Alfenas AC, Maffia LA, Ferreira EM, Binoti DHB, Mafia GMV (2009) Plant growth promoting rhizobacteria as agents in the biocontrol of eucalyptus mini-cutting rot. Trop Plant Pathol 34:10–17
Siddikee MA, Chauhan PS, Anandham R, Han G-H, Sa T (2010) Isolation, characterization, and use for plant growth promotion under salt stress of ACC deaminase-producing halotolerant bacteria derived from coastal soil. J Microbiol Biotechnol 20:1577–1584
Jaemsaeng R, Jantasuriyarat C, Thamchaipenet A (2018) Molecular interaction of 1-aminocyclopropane-1-carboxylate deaminase (ACCD)-producing endophytic Streptomyces sp. GMKU 336 towards salt-stress resistance of Oryza sativa L. cv. KDML105. Sci Rep 8:1950
Pierson LS, Pierson EA (2010) Metabolism and function of phenazines in bacteria: impacts on the behavior of bacteria in the environment and biotechnological processes. Appl Microbiol Biotechnol 86:1659–1670
de Vleesschauwer D, Cornelis P, Höfte M (2006) Redox-active pyocyanin secreted by Pseudomonas aeruginosa 7NSK2 triggers systemic resistance to Magnaporthe grisea but enhances Rhizoctonia solani susceptibility in rice. Mol Plant-Microbe Interact 19:1406–1419
Xu S, Pan X, Luo J, Wu J, Zhou Z, Liang X, He Y, Zhou M (2015) Effects of phenazine-1-carboxylic acid on the biology of the plant-pathogenic bacterium Xanthomonas oryzae pv. oryzae. Pestic Biochem Physiol 117:39–46
Das T, Ibugo AI, Klare W, Manefield M. Role of pyocyanin and extracellular DNA in facilitating Pseudomonas aeruginosa biofilm formation. In: Dhanasekaran D, Thajuddin N. Microbial biofilms—importance applications. IntechOpen; 2016. pp. 23–42
Kasim WA, Gaafar RM, Abou-Ali RM, Omar MN, Hewait HM (2016) Effect of biofilm forming plant growth promoting rhizobacteria on salinity tolerance in barley. Ann Agricult Sci 61:217–227
Berg G (2009) Plant-microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl Microbiol Biotechnol 84:11–18
Glick BR (2012) Plant growth-promoting bacteria: mechanisms and applications. Scientifica (Cairo) 2012:1–15
Semêdo LTAS, Linhares AA, Gomes RC, Manfio GP, Alviano CS, Linhares LF, Coelho RRR (2001) Isolation and characterization of actinomycetes from Brazilian tropical soils. Microbiol Res 155:291–299
Suela Silva M, Naves Sales A, Teixeira Magalhães-Guedes K, Ribeiro Dias D, Schwan RF (2013) Brazilian cerrado soil actinobacteria ecology. Biomed Res Int 2013:1–10
Acknowledgments
The authors thank Janaina Belquis da S. P. Langois and Rafaela Sole for their technical and laboratory assistance. The License for Research on Brazil’s Biodiversity was granted by the National Council for Scientific and Technological Development (CNPq 010539/2013-1).
Funding
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001, through fellowship of the first author and by the National Council for Scientific and Technological Development (CNPq/Brazil) through fellowship of the third author. Partial financial support was provided by CNPq/Brazil (403843/2013-8). In addition, this research was co-financed by Ballagro AgroTechnologia Ltda., São Paulo, Brazil (AGT/TA 01/2015-SIGPDI 194).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Responsible Editor: Luc F.M. Rouws
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Horstmann, J.L., Dias, M.P., Ortolan, F. et al. Streptomyces sp. CLV45 from Fabaceae rhizosphere benefits growth of soybean plants. Braz J Microbiol 51, 1861–1871 (2020). https://doi.org/10.1007/s42770-020-00301-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-020-00301-5