Abstract
Studies have established the pharmacological properties of Blighia sapida especially the glucose lowering potential without the elucidation of its mechanism of action. A negative regulatory action of protein tyrosine phosphatase 1B on insulin signaling pathway has identified the protein as a target for therapeutics in diabetes management. Therefore, this study aimed to investigate compounds from B. sapida with inhibitory potential against PTP 1B binding site. A library of compounds comprising of 56 compounds identified from B. sapida were screened against crystal structure of PTP 1B using glide docking model. The hit compounds were further subjected to molecular mechanics generalized born surface area (MM/GBSA) calculation to validate the binding energy; pharmacokinetics profile of the hit compounds was performed using maestro Schrodinger suite and the density functional theory (DFT) by Spatan 10. Based on molecular docking scores, molecular interaction with active catalytic residues and MM/GBSA calculation, eight (8) hit compounds were identified as potent inhibitors of PTP 1B compared with the co-crystalized inhibitor. Among the hit compounds, only 4,5-dicaffeoylquinic acid violated three Lipinski rules of five while other compounds obeyed the rule of five. The results of the frontier molecular orbitals revealed that the EHOMO values of the hit compounds range from − 6.07 to − 1.68 eV indicating that all the hit compounds will readily donate electron. The potential energy for the hit compounds range from − 180.468 to − 158.943 kJ/mol and 29.178 to 312.556 kJ/mol for low and high energy regions respectively. Conclusively, bioactive compounds from B. sapida may serve as promising therapeutic PTP 1B inhibitors for type-2 diabetes management.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Diabetes is a life-threatening disorder characterized by increase blood glucose level (hyperglycemia) and abnormal metabolism of carbohydrate, fat and protein with various degrees of polydipsia, polyuria and glycosuria [1]. Approximately 463 million people worldwide have been reported to be living with this pathogenic condition and projection of 24.8% increase in global diabetes prevalence by 2030 [2]. Diabetes pathogenesis are associated mainly with insulin dys-regulation which can be classified into two; damage to pancreatic β-cell resulting in decrease blood insulin levels in referred to as type-1-diabetes, while type-2 diabetes emanates from insensitivity or resistant to modulatory signals of plasma insulin [3]. Type-2 diabetes is capable of developing into various complications including cardiomyopathy, nephropathy, retinopathy, neuropathy etc. It is relative to some gene mutation, genetics, obesity etc [4].
The physiological function of insulin is elicited by its interaction with trans-membrane protein tyrosine kinase (PTK). These result in auto-phosphorylation of the protein and activation of insulin receptor substrate that propagates downstream insulin signaling processes [3]. Tyrosine phosphorylation and de-phosphorylation are essential processes in metabolic signaling pathways. Protein tyrosine kinase (PTK) causes phosphorylation while protein tyrosine phosphatases (PTPs) are responsible for de-phosphorylation thereby controlling cellular signaling [5]. Particularly, protein tyrosine phosphatase 1B (PTP 1B) has been linked to diabetes, it negatively regulates the insulin signaling pathway and its inhibition has been observed to show promising potential in the treatment of this life-threatening disease [5,6,7]. Therefore, PTP 1B is an established metabolic regulator and a pharmacological target for type-2 diabetes.
PTP 1B de-phosphorylates activated JAK 2 and STAT3, and prevent leptin signal transduction [8]. Increase PTP 1B expression influence the phosphorylation activity of PTK [9], which in turn resulted in failed insulin receptor and induced insulin resistance leading to type-2 diabetes and obesity [10]. Many PTP 1B inhibitors have been developed as therapeutic candidates for type-2 diabetes. This inhibitors including ertiprotafib, trodusquemine, thiazolidinediones, benzofuran, benzothiophene biphenyls are known to enhance insulin sensitivity in target tissues and improve glycemic control by ameliorating insulin resistance in both peripheral tissues and liver in type-2-diabetes [11]. Although these PTP 1B inhibitors exhibit good inhibitory efficacy, their clinical availability remains limited [12], coupled with the fact that majority of these synthetic pro-drugs were discontinued in phase II clinical trial due to their ineffectiveness and concentration-dependent adverse effects [3]. Thus, there is need to search for novel small molecules as selective inhibitor of PTP 1B for diabetes management.
Medicinal plants have contributed to the success of folklore medicine and account for over 60% of currently available anti-diabetes drug with few or no side effects including metformin isolated from Galega officinalis Linn plant [13]. According to Cragg and Newman [14], more than half of the approved drugs for treating metabolic disorders are plant derivatives. Blighia sapida Koenig, popularly known as Ackee is a medicinal plant commonly used as traditional herb among Nigerians [1]. The fruit has been reported to possess inhibitory effect against α-amylase and α-glucosidase [15]. The in-vitro antioxidant capacity and hepato-protective potential of B. sapida stem bark in STZ induced diabetes rats has been reported by Omoboyowa et al. [16] Saidu et al. [17] reported the blood glucose lowering potential of the root extract of B. sapida and the nephron-protective efficacy of B. sapida stem bark through down regulation of NGAL, KIM 1 and cystatin c in experimentally induced diabetes nephropathy has been reported by Omoboyowa et al. [1]. Herein, in silico model via molecular docking, MM/GBSA calculation, density function theory and pharmacokinetic study were employed to investigate the medicinal value of bioactive compounds from B. sapida as potent PTP 1B antagonist. This could serve as benchmark for developing PTP 1B therapeutic target for diabetes management.
2 Materials and Methods
Schrodinger suites software (version 2017-1) and Spartan 14 were used as computational tools in this present study.
2.1 Preparation of Protein Crystal Structure and Receptor Grid Generation
The X-ray crystallographic structures of Protein Tyrosin Phosphatase 1B (PTP 1B) was retrieved from the protein databank (https://www.rcsb.org/), with accession ID 2FJN. The protein preparation wizard of the Glide Schrödinger Suite 2017-1 was used to rectify specific errors in the protein during crystallographic structure. Missing hydrogen atoms in the protein structure were added, the bond orders were assigned and other options were used as default. Energetic optimization was performed in the final refinement step using OPLS3 force field and the RMSD of heavy atoms was set at 0.3 Å [18].
The glide grid file was generated via receptor grid generation panel of the Schrödinger Suite 2017–1 by picking the co-crystallized ligand (4-{2S, 4E)-2-(1H-1,2,3-benzotriazol-1-yl)-2-[4-(methoxycarbonyl)phenyl]-5-phenylpent-4-enyl}phenyl)(difluoro)methylphosphonic acid located at the active site of the protein to automatically generate x, y, z coordinates (36.71, 45.14 and 52.40 respectively). The Ramachandran plot of Protein Tyrosine Phosphatase 1B (PTP 1B) is illustrated in Fig. 1.
2.2 Natural Compounds Preparation
The structure-data file (SDF) structures of the fifty-six (56) compounds from Blighia sapida obtained from literature were downloaded from the PubChem database (https://pubchem.ncbi.nlm.nih.gov), imported on the workspace of Maestro Schrödinger suite Interface. Low energy 3D conformers with satisfactory bond lengths and angles for each two-dimensional structure were generated. The possible ionization states for each ligand structure were generated at a physiological pH of 7.2 ± 0.2. All other options were kept as default and the ligands were minimized using an optimized potential liquid simulation (OPLS3) force field [19].
2.3 Molecular Docking Studies
The screening of the fifty-six (56) compounds and co-crystalized inhibitor was carried out using three hierarchical GLIDE docking filtering, namely: high throughput virtual screening (HTVS), standard precision (SP) and extra precision (XP). HTVS docking is intended for the rapid screening of vast numbers of ligands. HTVS has much more restricted conformational sampling than SP docking, standard-precision (SP) docking is appropriate for screening ligands of unknown quality in large numbers. Extra-precision (XP) docking and scoring is a more robust and discriminating procedure, which takes longer to run than SP. XP is designed to be used on ligand poses that have been determined to be high-scoring using SP docking [18]. First, the fifty-six compounds were screened on the database through HTVS docking. The top eight (14.3%) scoring compounds were further docked using SP and XP, to score the compounds binding affinity via more expensive docking simulation on worthwhile poses. The co-crystallized ligand was re-docked into the catalytic site of 2FJN to confirm the accuracy of the screening and docking scores [20].
2.4 Prime MM/GBSA Calculations
Molecular mechanics generalized born surface area (MM/GBSA) calculation was carried out on the docking complexes, MM/GBSA is an advanced quantum mechanics calculation that helps to remove false-positive obtained from molecular docking analysis [21]. The docked ligands-PTP 1B complexes were minimized by using local optimization feature in Prime. The OPLS3 force field was employed to determine the binding energy (∆bind) for a set of ligands-PTP 1B complexes [21]. The following equation was used to calculate the binding free energy:
where ΔEMM: is the variance between the minimized energy of the ligands—PTP 1B complexes, ΔGSolv: is the variation between the GBSA solvation energy of the ligands—PTP 1B complexes and the sum of the solvation energies for the protein and ligand. In ΔGSA contains some of the surface area energies in the protein and ligand and the difference in the surface area energies for the ligands—PTP 1B complexes.
2.5 ADME/Tox
The pharmacokinetics profile necessary for predicting the drug-likeness property of the compounds was determined using Qikprop of Maestro Schrodinger suit (v11.12) [22].
2.6 Quantum Chemical Calculation
Quercetin, apigenin, luteolin, caffeic acid and chlorogenic acid were optimized at the DFT/Becke Three Lee Yang Parr/6-31G(d) level of theory [23] using Spartan 14 computational chemistry software. A restricted hybrid Hartree Fock-DFT self-consistent field calculation with Pulay’s direct inversion of the iterative sub-space and geometric direct minimization was employed [24]. The level of theory used was selected because it has been reported to be broadly consistent with experimental results [25] and useful in the interpretation of molecular mechanisms and interactions. The energies of the highest occupied molecular orbital (EHOMO), lowest unoccupied molecular orbital (ELUMO) and energy band gap, Eg (Eq. 1). The global reactivity descriptors were derived from the HOMO and LUMO energies (eqs. 2–5). Physicochemical parameters like molecular weight (MW), lipophilicity (log P), and polar surface area (PSA), hydrogen bond donor (HBD), hydrogen bond acceptor (HBA) were obtained in order to check for possible Lipinski violation(s) [26].
3 Results and Discussion
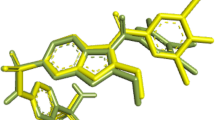
Before subjecting the B. sapida ligands to molecular docking study, the validation of the docking model was considered by extracting the co-crystalized ligand from the crystal structure, prepared and re-docked in the same active site. A root mean square deviation (RMSD) value of 1.04 Å was obtained upon super-imposition which was observed to be less than 2 Å, suggesting that the docking procedure is reproducible [27, 28]. As shown in Fig. 2, the co-crystalized ligand is green in colour and the re-docked ligand is red.
3.1 Molecular Docking Study and MM/GBSA Calculation
Molecular docking has been reported as an important model in the discovery and design of novel drug since it predicts the docking conformation and affinity of small molecules within the binding pocket of target proteins [29]. To predict potent therapeutic candidates from B. sapida bioactive compounds with high binding affinity and conformation against the binding pocket of PTP 1B, glide extra precision (XP) docking study and MM/GBSA calculation were carried out. The results of the molecular docking, post docking analysis and MM/GBSA calculation were presented in Table 1. The highest docking score corresponds to better binding affinity. Hence, isoferulic acid had the most favourable docking affinity against PTP 1B with binding energy of −8.575 kcal/mol. Next in the ranking were caffeic acid (−8.390 kcla/mol) and protocatechunic acid (−8.249 kcal/mol).
A total of eight (8) natural compounds from B. sapida emerged as promising inhibitor in terms of docking score which were comparable with the co-crystalized inhibitor (Fig. 3). Several studies have reported natural products as inhibitors of PTP 1B activity [3, 12] and docking affinity resonates the antagonistic prowess of ligands in protein–ligand complexes [30]. Hence, the results obtained in this study corroborates previous findings that the top docked natural compounds exhibit inhibitory potential against PTP 1B.
Interaction of natural compounds with the amino acid residues at the catalytic site of proteins is necessary for their inhibitory activity. The binding pocket of PTP 1B was observed to consist of essential residues including GLN 766, ALA 717, SER 716, CYS 715, ARG 721, GLY 720, ASP 548 which contributed to the PTP 1B-ligand interaction (Figs. 4, 5). Each ligand was critically analyzed for contact formation around the docked pose of PTP 1B at 4 Å distance, the results (Table 1; Fig. 4) showed that isoferulic acid and caffeic acid formed eight (8) H-bonds with the amino acid residues at the catalytic site and protocatechunic acid formed six (6) H-bonds. All the B. sapida ligands formed more H-bonds compared with the co-crystalized inhibitor with two (2) H-bonds interaction with ASP 548 and ARG 524 residues. The formation of H-bond interaction between the natural ligands and PTP 1B binding site might result from the functional groups present in the ligand structure for instance, the three top scored ligands with highest number of H-bond formation i.e. isoferulic acid (8 H-bonds), caffeic acid (8 H-bonds) and protocatechunic acid (6 H-bonds) possess carboxyl group (COO−) which formed seven H-bonds with the hydrophobic amino acid residues while the extra H-bond formed in isoferulic acid and caffeic acid is between the hydroxyl group on the phenyl ring and GLN 762 of the active site of PTP 1B. Therefore, the interaction of the ligands’ carboxyl group with the amino acid residues which lead to strong H-bond interaction may be responsible for the favoured binding energy with PTP 1B and contribute to the stability of the compounds with the protein. The result obtained in this study is consistent with the finding of Olawale et al. [3] who reported favoured binding energy of natural compounds resulting from H-bond interaction between the carbonyl group of the compounds and the amino acid residues of PTP 1B.
Although molecular docking is widely used for computer aided drug design (CADD), the model has been reported to be deficient of some energy parameters including solvation energy system. Hence, the validation procedure was further supported by prime MM/GBSA calculation. MM/GBSA has been reported for its reliability and accuracy in computing structural stability and binding energy of protein–ligand complexes [28]. A positive binding free energy score (MM/GBSA) score depicts false binding energy (docking score). Remarkably, the results of the binding free energy obtained from the investigated natural compounds and inhibitor formed favorable stability with PTP 1B with negative binding free energy values (Table 1; Fig. 3). PTP 1B-isoferulic acid (-53.393) and PTP 1B-caffeic acid (− 52.430) complexes were computed with the highest binding free energy among the ligands and the PTP IB-inhibitor complex (− 31.214) was observed to give the lowest binding free energy.
Inhibitor: (4-{2S, 4E)-2-(1H-1,2,3-benzotriazol-1-yl)-2-[4-(methoxycarbonyl)phenyl]-5-phenylpent-4-enyl}phenyl)(difluoro)methylphosphonic acid.
3.2 ADMET Profile of Hit Compounds
The evaluation of pharmacokinetic profile of drug candidates is important in the discovery and design of novel therapeutic agents. Thus, significant failure rate of potential drug at late stage of development have been attributed to poor absorption, distribution, metabolism and excretion with high toxicity rate [31]. Therefore, in silico model remains the fastest and cost-effective techniques of ADMET screening. Here in, certain pharmacokinetic properties of the hit compounds such as madin-Darby canine kidney (MDCK) cell, apparent calcium carbonate (QPCaco), IC50 value for the blockage of HERG K (QPPHERG), the binding to human serum albumin (QPlogkhsa) and brain/blood partition coefficient (QPlogBB) were predicted by Qikprop tool of Schrodinger suite.
The results of the pharmacokinetic properties shown in Table 2 revealed that, cimicfugic acid D, 4,5-dicaffeoylquinic acid and Gallic acid exhibit poor Caco cell permeability below 25 nm/s. The Caco cell lines are usually use as a model of human intestinal absorption of drug candidates [32]. Cimicfugic acid D, 4,5-dicaffeoylquinic acid and ferulic acid were also observed to be below the normal range of brain/blood partition coefficient (− 3.0 to 1.2). This brain/blood partition coefficient is crucial in the assessment of leading drug candidates and MDCK cells are involved in drug development to investigate the rate of drug efflux, predict passive permeability and active transport [33]. All other compounds fall within the recommended range of the pharmacokinetic parameters with good percentage oral absorption but 4,5-dicaffeoylquinic acid and cimifugic acid D were observed to have low percentage oral absorption.
The drug likeness prediction of the hit compounds was presented in Table 3, except 4,5-dicaffeoylquinic acid with molecular weight of 516.457 Da, all other compounds fall within the recommended range for molecular weight. Only cimicfugic acid D and 4,5-dicaffeoylquinic acid exhibit a high polar surface area of 206.775 and 238.863 respectively. Other compounds fall within the normal range (7–200). Several rules have been use to evaluate the drug likeness of small molecules, with most popular being the Lipinski rule of five (RO5) [18]. The rules (MW < 500DA, H-bond donor ≤ 5, H-bond acceptor ≤ 10 and octanol–water partition coefficient < 5) [34] states that, a drug-like molecule must not violate more than one of the rules of five [35]. Results from this study (Table 3) showed that all the hit compounds except 4,5-dicaffeoylquinic acid (violate 3) obeyed Lipinski rules. Therefore, they can be predicted as potential drug candidates.
3.3 Frontier Molecular Orbitals
The high occupied molecular orbital (HOMO) and low unoccupied molecular orbital (LUMO)energies as well as other reactivity descriptor from density function theory calculations are utilize to predict the reactivity of small molecule. Thus, frontier molecular orbitals (FMOs) describe the interaction and overlapping of molecules to provide information about the transfer of electron in a molecular and chemical reactivity and stability of the molecules [35]. The HOMO energy (EHOMO) predicts the electron donating capacity of a molecule while the LUMO energy (ELUMO) predicts the electron accepting ability of a molecule. Hence, higher value of EHOMO and lower value of ELUMO are responsible for the increase reactivity of molecules [36]. Results from this study (Fig. S1 and Table 4) revealed that the EHOMO values of the hit compounds range from − 6.07 to − 1.68 eV indicating that all the hit compounds will readily donate electron while the ELUMO values ranges from − 1.63 to − 1.06 eV. The result agrees with the report of Oyeneyin et al. [36]. The energy band gaps (Eg) were calculated to vary from 4.19 to 5.00 eV suggesting the reactivity and stability of the compounds, since Eg has been reported as an indicator of the chemical reactivity and stability of a molecule with higher Eg value denotes greater stability, low reactivity and less bioavailability [37].
Further investigation into the stability and reactivity of the compounds was obtained by calculating the global reactivity descriptor using chemical hardness (η), softness (δ), electronegativity (χ) and chemical potential (Cp) as studied parameters. From Table 4, the chemical hardness and softness values for the hit compounds range from 2.05 to 2.50 eV and 0.40 to 0.49 eV−1 respectively with ferulic acid having the lowest hardness and highest softness values among the hit compounds. According to Pearson [38], the high chemical hardness and low softness values of a molecule translate to higher stability and lower reactivity. Therefore, ferulic acid is predicted as the compound with the highest stability and lowest reactivity compounds among the hits. Electronegativity (χ) predicts the ability of a molecule to attract electron [39]. The electronegativity values for the hit compounds range between 3.53 to 3.18 eV, with P-coumaric acid (3.81 eV) having the highest value, Thus highest ability for attracting electron towards itself.
The molecular electrostatic potential (MEP) gives insight into the part of the compounds which is electron rich and electron deficient. It predicts the reactive sites on molecules where electrophilic and nucleophilic interactions are possible [37, 40]. From Fig. 6, the red, green and blue colours on the map depict increasing order of electrostatic potential as negative, zero and positive. The negative (red) regions are low energy sites for electrophilic interaction by acids, located close to the oxygen atom within the compounds. The blue (positive potential) regions are high energy site which are located on the hydrogen atom attached to nitrogen atoms therefore repels proton due to extremely low electron density around it. This region is for nucleophile interaction by bases. The potential energy for the hit compounds range from − 180.468 to − 158.943 kJ/mol for low energy regions and 2–9.178 to 312.556 kJ/mol for high energy regions. The presence functional groups such as carbonyl group, carboxyl group and hydroxyl group on the structure of the natural compounds imposes asymmetric charge distribution and creates nucleophilic and electrophilic sites for effective interaction with the PTP 1B active site. This suggests that the hit compounds could be predicted as good ligands.
4 Conclusion
Fifty-six bioactive compounds of B. sapida were screened for potential inhibitory activity against PTP 1B, a therapeutic target in the management of diabetes. Based on the molecular docking and post docking analysis, eight (8) hit ompounds were identified as potent inhibitors of PTP 1B compared with the co-crystalized inhibitor. All eight hit compounds possess favourable ADMET profile and only 4,5-dicaffeoylquinic acid violated more one Lipinski rule of five. These hit compounds are capable nucleophilic and electrophilic interactions with the active site of PTP 1B receptor. Hence, the results from this in silico study predict eight (8) natural compounds previously identified from Blighia sapida plant as potent inhibitors of protein tyrosine phosphatase 1B (PTP 1B). These compounds are therefore propose as lead compounds and should be explore in further in vivo and in vitro experiment for the design of PTP 1B inhibitors in the therapeutic management of diabetes and its associated complication.
Availability of Data and Materials
Data were supplied in the supplementary file.
Code Availability
Not applicable.
References
Omoboyowa DA, Karigidi KO, Aribigbola TC (2021) Nephro-protective efficacy of Blighia sapida stem bark ether fractions on experimentally induced diabetes nephropathy. Comp Clin Pathol. https://doi.org/10.1007/s00580-020-03186-w
Williams R, Colagiuri S, Almutairi R, Montoya PA, Basit A, Beran D, Besançon S, Bommer C, Borgnakke W, Boyko E (2019) IDF diabetes atlas. Int diabetes federation Press, UK, pp 12–15
Olawale F, Olofinsan K, Iwaloye O, Chukwuemekae PO, Elekofehinti OO (2021) Screening of compounds from Nigerian antidiabetic plants as protein tyrosine phosphatase 1B inhibitor. Comput Toxicol. 21:100200
Halim M, Halim A (2019) The effects of inflammation, aging and oxidative stress on the pathogenesis of diabetes mellitus (type 2 diabetes). Diabetes Metab Syndr 13(2):1165–1172. https://doi.org/10.1016/j.dsx.2019.01.040
Figueiredo A, Leala EC, Carvalho E (2020) Protein tyrosine phosphatase 1B inhibition as a potential therapeutic target for chronic wounds in diabetes. Pharm Res 159:104977
Shi K, Egawa K, Maegawa H, Nakamura T, Ugi S, Nisho Y, Kashiwagi A (2004) Protein-tyrosine phosphatase 1B associates with insulin receptor and negatively regulates insulin signaling without receptor internalization. J Biochem 136(1):89–96. https://doi.org/10.1093/jb/mvh094
Liu ZQ, Liu T, Chen C, Li M, Wang Z, Chen R, Wei G, Wang X, Luo D (2015) Fumosorinone, a novel PTP1B inhibitor, activates insulin signaling in insulin-resistance HepG2 cells and shows anti-diabetic effect in diabetic KKAy mice. Toxicol Appl Pharmacol 285(1):61–70. https://doi.org/10.1016/j.taap.2015.03.011
Lund IK, Hansen JA, Andersen HS (2005) Mechanism of protein tyrosine phosphatase 1B-mediated inhibition of leptin signalling. J Mol Endocrinol 34(2):339–351
He RJ, Yu ZH, Zhang RY (2014) Protein tyrosine phosphatases as potential therapeutic targets. Acta Pharmacol Sin 35(10):1227–1246
Barr AJ (2010) Protein tyrosine phosphatases as drug targets: strategies and challenges of inhibitor development. Future Med Chem 2(10):1563–1576
Sun QC, Wang Y, Huang H, Zhang M, Zou W (2016) Type 2 diabetes mellitus and protein-tyrosine phosphatase 1B. J Diabetes Metab Disord Control. 3(8):180–183. https://doi.org/10.15406/jdmdc.2016.03.00096
Zhao BT, Nguyen DH, Le DD, Choi JS, Min BS, Woo MH (2018) Protein tyrosine phosphatase 1B inhibitors from natural sources. Arch Pharm Res 41(2):130–161. https://doi.org/10.1007/s12272-017-0997-8
Patade G, Marita A (2014) Metformin: a journey from countryside to the bedside. J Obes Metab Res 1:127
Cragg GM, Newman DJ (2013) Natural products: a continuing source of novel drug leads. Biochim Biophys Acta 1830(6):3670–3695
Kazeem MI, Ogungbe SM, Saibu GM, Aboyade OM (2014) In vitro study on the hypoglycaemic potential of Nicotiana tabacum leaf extracts. Bangladesh J Pharmacol 9:140–145
Omoboyowa DA, Akintimehin ES, Akinnubi CO, Balogun TA (2019) In vitro antioxidant and hepato-protective potential of Blighia sapida stem bark ether fractions in STZ induced diabetes rats. J Herbal Drug 4(10):139–146
Saidu AN, Mann A, Onuegbu CD (2012) Phytochemical screening and of aqueous Blighia sapida root bark extract on normoglycemic albino rats. Br J Pharm Res 2:89–97
Elekofehinti OO, Iwaloye O, Josiah SS, Lawal AO, Akinjiyan MO, Ariyo EO (2020) Molecular docking studies, molecular dynamics and ADME/tox reveal therapeutic potentials of STOCK1N-69160 against papain-like protease of SARS-CoV-2. Mol diver 25:1761–1773
Harder E, Damm W, Maple J, Wu C, Reboul M, Xiang JY (2015) OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J Chem Theory Comput 12(1):281–296. https://doi.org/10.1021/acs.jctc.5b00864
Akinloye OA, Akinloye DI, Onigbinde SB, Metibemu DS (2020) Phytosterols demonstrate selective inhibition of COX-2: in-vivo and in-silico studies of Nicotiana tabacum. Bioorg Chem 102:104037
Omoboyowa DA (2021) Sterols from Jatropha tanjorensis leaves exhibit anti-inflammatory potential: in vitro and in silico studies. Bull Natl Res Centre 45:194–207
Schrödinger Release 201 8-4 (2017) Schrödinger. LLC, New York. 2017-1
Jensen F (2001) Polarization consistent basis sets: principles. J Chem Phys 115:9113–9125
Becke AD (1993) Becke’s three parameter hybrid method using the LYP correlation functional. J Chem Phys 98:5648–5652
Odewole OA, Ibeji CU, Oluwasola HO, Oyeneyin OE, Akpomie KG, Ugwu CM, Ugwu CG, Bakare TE (2021) Synthesis and anti-corrosive potential of Schiff bases derived 4-nitrocinnamaldehyde for mild steel in HCl medium: experimental and DFT studies. J mol. Struct 1223:1–9. https://doi.org/10.1016/j.molstruc.2020.129214
Adejoro I, Ibeji C, Akintayo D (2017) Quantum descriptors and corrosion inhibition potentials of Amodaquine and Nivaquine. Chem Sci J 8:149
Kamchonwongpaisan S, Quarrell R, Charoensetakul N, Ponsinet R, Vilaivan T, Vanichtanankul J (2004) Inhibitors of multiple mutants of Plasmodium falciparum dihydrofolate reductase and their antimalarial activities. J Med Chem 47(3):673–680
Balogun TA, Iqbal MN, Saibu OA, Akintubosun MO, Lateef OM, Nneka UC, Abdullateef OT, Omoboyowa DA (2021) Discovery of potential HER2 inhibitors from Mangifera indica for the treatment of HER2-positive breast cancer: an integrated computational approach. J Biomol Struct Dyn 39:1–12
Mielech AM, Chen Y, Mesecar AD, Baker SC (2014) Nidovirus papain-like proteases: multifunctional enzymes with protease, deubiquitinating and deISGylating activities. Virus Res 194:184–190
Cho JK, Curtis-Long MJ, Lee KH, Kim DW, Ryu HW, Yuk HJ, Park K (2014) Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia tomentosa. Bioorg Med Chem 21:3051–3057
Omoboyowa DA, Omomule OM, Balogun TA, Saibu OA, Metibemu DS (2021) Protective potential of ethylacetate extract of Abrus precatorius (Linn) seeds against HCl/EtOH-induced gastric ulcer via pro-inflammatory regulation: In vivo and in silico study. Phytomed Plus 1:100145
Van-Breemen RB, Li Y (2005) Caco-2 cell permeability assays to measure drug absorption. Expert Opin Drug Metab Toxicol 1(2):175–185
Jin X, Luong TL, Reese N, Gaona H, Collazo-Velez V, Vuong C (2014) Comparison of MDCK-MDR1 and Caco-2 cell based permeability assays for anti-malarial drug screening and drug investigations. J Pharmacol Toxicol Methods 70(2):188–194
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46:3–26. https://doi.org/10.1016/s0169-409x(00)00129-0
Balogun TA, Ipinloju N, Abdullateef OT, Moses SI, Omoboyowa DA, James AC, Saibu OA, Akinyemi WF, Oni EA (2021) Computational evaluation of bioactive compounds from Colocasia affinis Schott as a novel EGFR inhibitor for cancer treatment. Cancer Inform 20:1–12
Oyeneyin OE, Abayomi TG, Ipinloju N, Agbaffa EB, Akerele DD, Arobadade OA (2021) Investigation of amino chalcone derivatives as anti-proliferative agents against MCF-7 breast cancer cell lines-DFT, molecular docking and pharmacokinetics studies. Adv J Chem A 4(4):288–299. https://doi.org/10.22034/AJCA.2021.285869.1261
Uzzaman M, Mahmud T (2020) Structural modification of aspirin to design a new potential cyclooxygenase (COC-2) inhibitors. In Silico Pharmacol 8:1
Pearson RG (1993) The principle of maximum hardness. Acci Chem Res 26:250–255
Geerlings P, De-Proft F (2002) Chemical reactivity as described by quantum chemical methods. Int J Mol Sci 3:276–306
Noureddine O, Issaoui N, Al-Dossary O (2021) Quantum chemical calculations, spectroscopic properties of a novel piperazine derivatives. J King Saud Univ Sci 33:101248
Acknowledgements
The author sincerely acknowledges Dr. Oyeneyin E. O. of the Department of Chemical Science, Adekunle Ajasin University for providing the Spatan 10 software utilized in the DFT analysis.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
DAO performed all the computational analysis and wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The author declares no competing interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Omoboyowa, D.A. Virtual Screening of Phyto-compounds from Blighia sapida as Protein Tyrosine Phosphatase 1B Inhibitor: A Computational Approach Against Diabetes. Chemistry Africa 5, 871–881 (2022). https://doi.org/10.1007/s42250-022-00373-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42250-022-00373-w