Abstract
Detection and characterization of changes in virulence patterns of pathogen populations is a prelude to effective management of wheat rusts. Such information is useful for the identification of sources of resistance, pre-emptive breeding and judicious deployment of resistant wheat cultivars. During 2013–14, three new Yr9-virulent pathotypes (110S119, 238S119 and 110S84) of Puccinia striiformis f. sp. tritici (Pst) possessing combined virulence to cvs Strubes Dickkopf (Yr2, Yr3a, Yr4a) and Suwon92xOmar (YrSU) were detected in India. These pathotypes were more aggressive and virulent than those reported earlier. Riebesel 47/51 (Yr2, Yr9) and Hobbit (Yr14) hitherto resistant to Indian Pst populations, became susceptible to 238S119 and 110S119, respectively. Moreover, the bread wheat cv. HD2967, currently occupying a 10–12 million ha area, showed a high level of susceptibility to these pathotypes. Stripe rust resistance genes, Yr1, Yr5, Yr10, Yr15, Yr24, Yr28 and YrSP were found effective against the new pathotypes. Phylogenetic relationships of the new and nine other closely related pathotypes were elucidated by comparing sequences of the internal transcribed spacer regions. It appeared that pathotypes110S84 and 110S119 were distinct and evolved independently, whereas 238S119 could have evolved from 46S119 by gaining virulence to cvs Strubes Dickkopf and Riebesel 47/51. Recently developed Indian bread wheat germplasm comprising 56 newly released cultivars and 64 advanced lines (2016–17) were evaluated under controlled conditions at the seedling stage for resistance to the three new and the two already known Yr9-virulent pathotypes 46S119 and 78S84. None of the cultivars was resistant to the new pathotypes. Cultivars DBW88, DPW621–50, HD3043, HD3059, HD3171, HS507, HS542, HS562, MACS6478, PBW723, WH1021 and WH1105, resistant to the erstwhile prevalent pathotypes 46S119 and 78S84, showed susceptibility to the new pathotypes. However, 11 advanced bread wheat lines viz. DBW246, HS645, PBW752, PBW757, PBW777, PBW779, PBW780, UP2993, VL1012, VL3013 and WH1233 were found resistant to all the test pathotypes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Stripe (Yellow) rust caused by Puccinia striiformis f. sp. tritici, is a devastating disease of wheat. It occurs frequently in the cooler wheat-growing areas with moist weather conditions (Chen et al. 2014). In India, the disease primarily occurs in the North Western Plains Zone (NWPZ) and Northern Hills Zone (NHZ). Stripe rust potentially threatened the Indian wheat production in 1982 when it appeared in epidemic form in Punjab, Haryana, Western Uttar Pradesh and Jammu & Kashmir States (Nagarajan et al. 1984). Stripe rust can cause 100% yield loss under conducive environment, however, crop damage remains generally in the range of 10–70% depending upon growth stage, rust severity, duration of infection and susceptibility of cultivars (Chen and Penman 2005). Yield losses in wheat due to stripe rust infection are usually ascribed to reduction of kernel numbers per spike, low test weight, and reduced kernel quality due to decreased photosynthesis (Prescott et al. 1986).
Reports of severe stripe rust infection have historically been associated with the appearance of new Pst races which could overcome the deployed host resistance (Nagarajan and Joshi 1975; Nayar et al. 1997; Prashar et al. 2007). Hence, continuous monitoring of Pst population is necessary for managing wheat stripe rust through the development and deployment of resistant cultivars. The 1BL/1RS translocation lines have been widely used in wheat breeding for their resistance to all the three rust and powdery mildew (Sr31/Lr26/Yr9/Pm8), high yield potential and tolerance to abiotic stresses (Zeller et al. 1973). In India, a number of wheat cultivars CPAN3004, PBW175, PBW343, PBW373, UP2338, UP2382, UP2418 and UP2425 carrying 1BL/1RS translocation (Kumar et al. 2003) were released during the 1990s for commercial cultivation. As anticipated, two Yr9-virulent pathotypes, 46S119 and 78S84, were detected in 1996 and 2001, respectively (Prashar et al. 2007). Subsequently, three additional Yr9-virulent pathotypes, 110S119, 238S119, and 110S84 possessing combined virulence to cvs Strubes Dickkopf (Yr2, Yr3a, Yr4a) and Suwon 92 x Omar (YrSU), were detected during 2013–14. These pathotypes were more aggressive and virulent than those reported earlier. Riebesel 47/51 (Yr2, Yr9) and Hobbit (Yr14) hitherto resistant to Indian Pst populations, became susceptible to 238S119 and 110S119, respectively. Phylogenetic relationships of these new pathotypes with nine other closely related pathotypes were studied using host differentials and internal transcribed spacer (ITS) regions. White et al. (1990) designed universal primer pair ITS 1 and ITS 4 which amplifies the ITS region (600–800 bp) and is considered appropriate for determining phylogenetic relationships (Lee and Taylor 1992; Cooke and Duncan 1997).
In India, the fungicides propiconazole 25% EC (tilt), tebuconazole 25% EC (folicur) and triadimefon 25% EC (Bayleton) are usually recommended at the rate of 0.1% (Bhardwaj et al. 2016), as an emergent tool for managing wheat rusts under high disease incidence. However, genetic resistance has been the most effective, economic and eco-friendly approach for the management of wheat rust diseases. A dynamic rust resistance breeding programme always thinks ahead of the pathogen through identification of sources of resistance to any new pathotype in addition to the prevalent ones. Recently developed Indian bread wheat germplasm, comprising 56 newly released cultivars (2000–2016) and 64 advanced lines (2016–17), was evaluated under controlled conditions at the seedling stage for resistance to the three new and two previously reported Yr9-virulent pathotypes 46S119 and 78S84. Our findings related to characterization of the new Pst pathotypes and identification of resistance sources are reported in the present communication.
Materials and methods
Analysis of rust samples and identification of new pathotypes
Stripe rust-infected samples were collected from farmers fields in Northern India. Two-to-three bits of leaf tissue (3–4 cm) from each sample were placed in Petri plates containing 2% water agar and incubated at 4 °C for 48 h for inducing sporulation. One-week-old seedlings of the susceptible durum wheat line A-9-30-1 were inoculated with fresh urediniospores from each sample and, thereafter, incubated in a moist chamber (RH >80%) at 12 ± 2 °C for 48 h. Subsequently, these plants were transferred onto greenhouse benches where 16 ± 2 °C temperature, 60–80% relative humidity and 15,000 lx illumination for 12 h period were maintained. To prevent contamination, plants of different samples were separated by rectangular screens made of a wooden frame with muslin cloth. Fresh urediniospores collected from these plants 15–20 days post inoculation (dpi), were used to inoculate the sets of wheat differentials with a lancet needle (Nayar et al. 1997).
The Indian system of pathotype identification and nomenclature is based on three sets of differentials. Set-A is similar to world differentials and set-B comprises selected Indian and European differentials (Table 1). Set-0 is supplemental, helping to reveal minor differences in pathogenicity. Seedlings of differentials were grown in aluminum bread pans (29 cm long × 12 cm wide × 7 cm deep), containing autoclaved (60 °C for 1 h) mixture of fine loam and farmyard manure (3:1). Seven- to eight-day-old seedlings of differentials were inoculated with fresh urediniospores and sprayed with a fine mist of water following the aforementioned procedures.
Infection types of Pst isolates on stripe rust differentials were scored at 16 dpi following McIntosh et al. (1995). Infection types were characterized as 0 (Immune) = no visible uredia; 0; and; (very resistant) = necrotic flecks; N (resistant) = necrotic area without sporulation; 1 (resistant) = necrotic and chlorotic areas with restricted sporulation; 2 (moderately resistant) = moderate sporulation with necrosis and chlorosis; 3 (moderately susceptible) = sporulation with chlorosis, 3+/4 (susceptible) = abundant sporulation without chlorosis. Infection type 33+ was classified when both 3 and 3+ pustules occurred together. If a Pst isolate produced infection types on differentials dissimilar from those of the already reported pathotypes, isolations were made and used for further inoculations. Infection types were reconfirmed and new pathotypes were designated in accordance with the binary notation system used in India (Nagarajan et al. 1985; Bhardwaj et al. 2012).
Identification of resistance sources among bread wheat germplasms against new pathotypes
For identifying the sources of resistance, 56 newly released cultivars and 64 advanced lines were evaluated against three new (110S119, 238S119, 110S84), and two earlier detected Yr9-virulent Pst pathotypes (46S119 and 78S84). The response of 21 known Yr-genes to these pathotypes was also assessed. Near-isogenic lines (NILs) in Avocet background were the source of Yr-genes except cv. Hobbit (Yr14). Wheat lines were grown in aluminum bread pans in a mixture of fine loam and farmyard manure (3:1). These trays were sufficiently large to accommodate 18 wheat lines and a susceptible control. For each wheat line, 5 to 6 seeds were sown in hills. One-week-old seedlings were inoculated using a glass atomizer containing 10 mg spores of a Pst pathotype suspended in 2 ml light grade mineral oil (Soltrol 170; Chevron Phillips Chemicals Asia Pte. Ltd., Singapore). After inoculation, the oil was allowed to evaporate for 5 min. The same procedure mentioned above was followed for differentials and susceptible hosts. The experiment was repeated twice to ascertain the infection types.
Molecular characterization of new pathotypes
The genetic variability among Pst pathotypes was explored by comparing the sequences of the internal transcribed spacer regions (ITS) of nuclear ribosomal DNA. Genomic DNA was isolated from dried urediniospores using the cetyletrimethylammonium bromide (CTAB) method as previously described (Kiran et al. 2017). ITS regions, including the ribosomal 5.8S RNA gene, were amplified using primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) that annealed to the flanking 18S and 28S rRNA (White et al. 1990). The PCR reaction was carried out in a 25-μl reaction mixture containing the following components: 10× PCR buffer, 50 ng DNA template, 2 mM MgCl2, 0.25 mM dNTP mixture and 0.25 μM each of primer, and one unit of Taq polymerase (Bangalore Genei, India). Amplifications were performed in Thermal Cycler (Applied Biosystems, USA) under the following conditions: initial denaturation 5 min at 94 °C, 35 cycles of 45 s at 94 °C, 45 s at 52 °C, 1 min at 72 °C, with the final extension of 10 min at 72 °C. The PCR product (600 bp) was purified using Wizard Genomic DNA Purification Kit (Promega, USA) and sequenced using BigDye TM Terminator v.3.1 chemistry (Applied Biosystems, USA) on an ABI Prism TM310 Genetic Analyzer (Applied Biosystems, USA). The ITS sequences of pathotypes were aligned using the ClustalW (Larkin et al. 2007) against the corresponding nucleotide sequences retrieved from GenBank. Using MEGA7 (Kumar et al. 2016), the best-fitting nucleic acid substitution was calculated in maximum likelihood (ML) model test. The model with lowest Bayesian Information Criterion (BIC) score was selected and used for further phylogenetic analysis using MEGA7 (Kumar et al. 2016), which includes distance matrix construction and phylogeny reconstruction using ML with a Tamura 3-parameter model (Tamura 1992).
Results
Pathological characterization and confirmation of new pathotypes
A total of 321 stripe rust-infected samples of bread wheat were analyzed on Indian and European rust differentials during 2013–14. Three new Pst pathotypes were identified in these samples in addition to the six reported earlier. A stripe rust sample, collected from the wheat cv. WL711 (pedigree-S308/CHR//KAL) from the Ropar district of Punjab, was similar to pathotype 46S119 but had additional virulence for Suwon92xOmar (YrSU) (Table 2). This isolate, designated as 110S119 (110E159), carried unique virulence for both cvs Strubes Dickkopf and Suwon92xOmar.
Similarly, another stripe rust sample, collected from the wheat cultivar RAJ3777 (RAJ3160/HD2449) in 2014 from Sirmaur district of Himachal Pradesh, had infection types similar to those of the pathotype 78S84, except that it possessed virulence for cv. Strubes Dickkopf (Yr2, Yr3a, Yr4a). Isolations made from cvs Strubes Dickkopf and Suwon92xOmar confirmed their virulence for both these differentials based on reinoculation. Thus, this isolate was different from the pathotypes earlier reported from India, and was designated as 110S84 (110E22).
In India, cv. Riebesel-47/51 was resistant to known Pst pathotypes until 2014, when a stripe rust sample collected from the wheat cultivar HS507 (KAUZ/MYNA/VUL//BUC/FLK/4/ MILAN) from Bilaspur district of Himachal Pradesh showed virulence for it. Isolations were made from cvs Strubes Dickkopf, Suwon92xOmar and Riebesel-47/51 and simultaneously compared with the closely related pathotypes 46S119 and 78S84. All the three test isolates were found possessing virulence for cv. Riebesel-47/51 along with combined virulence for cvs Strubes Dickkopf and Suwon92xOmar. The new pathotype was designated as 238S119 (238E159). The avirulence/virulence formulae of new pathotypes are given in Table 2.
Differentiating characteristics of new Yr9-virulences in comparison with the related pathotypes are shown in Table 3. It appears that the new pathotype 110S119 evolved by single step forward point mutation in pathotype 46S119, resulting in additional virulence on cv. Suwon92xOmar (YrSU). Similarly, pathotype 110S84 could have evolved from 78S84 by acquiring virulence to cv. Strubes Dickkopf (Yr2, Yr3a, Yr4a). Although pathotypes 110S119 and 110S84 possessed combined virulence for cvs Strubes Dickkopf and Suwon92xOmar, they can be differentiated on cvs Hybrid46 (Yr3b, Yr4, Yr4b), Heines VII (Yr2, Yr11, Yr25, +) and Sonalika (Yr 2, YrA). The former (110S119) is virulent to these differentials, while the latter (110S84) is avirulent. The gene YrA (cv. Sonalika) showed incompatible interaction with 110S84, but compatible with 110S119 and 238S119. The pathotype 238S119 was unique as it possessed additional virulence to cv. Riebesel-47/51(Yr2, Yr9, +), to which the other pathotypes were avirulent.
Molecular characterization of new pathotypes
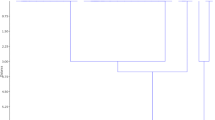
Phylogenetic relationships among 12 Pst pathotypes were studied by comparing their ITS sequences (Table 4). DNA sequences from Pst pathotypes were clustered into two major clades by using the Maximum Likelihood method based on the Tamura 3-parameter model (Fig. 1). Interestingly, all the Yr9-avirulent pathotypes (47S102, 66S64–1, 66S0, 67S64, 70S4, 70S0–2 and 38S102) were grouped in clade I, and the Yr9-virulent ones (46S119, 238S119, 110S119, 78S84 and 110S84), in clade II. Pathotypes 46S119 and 238S119 in clade II, showed close similarity as suggested by bootstrap value of 63%. It appears that the pathotype 238S119 evolved from 46S119 by gaining virulence to cvs Suwon92xOmar and Riebesel 47/51. Other two pathotypes, 110S119 and 110S84 were distinct from each other and could have evolved independently. Similarly, in clade I, pathotype 70S4 had close relationship with pathotype 67S64, as based on bootstrap value of 65%.
Phylogenetic tree based on comparisons of ITS-rDNA sequences of new Pst pathotypes and sequences of related pathotypes from GenBank, constructed by using the Maximum Likelihood method based on the Tamura 3-parameter model. The percentage of trees in which the associated taxa clustered together is shown next to the branches. * new pathotype
Sources of resistance in bread wheat germplasms
A collection of 56 recently released bread wheat cultivars (released by the Central Variety Release Committee during 2000–2016) and 64 advanced lines were seedling-evaluated for resistance to the new and earlier reported Yr9-virulences (110S119, 238S119, 110S84, 46S119 and 78S84). In all, 25 and 41% of the cultivars were resistant to 46S119 and 78S84, respectively. Cultivars HS 507 (1.79% of the total cultivars evaluated); HD 3043 and PBW 723 (3.57%); and DBW39, HD2733, HD3043, HD3171, HS507, KRL213 and PBW723 (14.28%) showed resistance to 110S119, 238S119, and 110S84, respectively (Fig. 2). None of the released cultivars was resistant to all the five pathotypes. Cultivars DBW88, DPW621–50, HD3043, HD3059, HD3171, HS507, HS542, HS562, MACS6478, PBW723, WH1021 and WH1105, hitherto resistant to Yr9-virulent pathotypes, 46S119 and 78S84, showed susceptibility to the new pathotypes (Supplementary Table S1). In advanced bread wheat lines, 11 (DBW246, HS645, PBW752, PBW757, PBW777, PBW779, PBW780, UP2993, VL1012, VL3013 and WH1233) were resistant to all pathotypes. However, 14 lines (CG1023, DBW179, DBW189, HI1617, HPW440, HPW448, HPW449, HS630, HS644, HS647, MP1318, UP2942, VL3015 and WH1316), resistant to pathotypes 46S119 and 78S84, showed susceptibility to the new pathotypes (Supplementary Table S2). Among the advanced lines, 20.31, 20.31 and 59.38%, were found resistant, intermediate and susceptible, respectively, to pathotype 110S119; while to pathotype 110S84, 65.63 and 12.5% lines were resistant and susceptible, respectively (Fig. 3).
Among the 21 Yr-genes, Yr1, Yr5, Yr10, Yr15, Yr24, Yr28 and YrSP were effective against all the five Yr9-virulent pathotypes. Gene Yr14 was defeated by new virulent pathotype 110S119. Genes Yr11, Yr17, Yr18, Yr29 and YrA were resistant to pathotypes 78S84 and 110S84, but susceptible to pathotypes 46S119, 110S119 and 238S119 (Table 5).
Discussion
In India, Yr9-virulences appeared primarily due to extensive deployment of 1BL/1RS translocation lines. Popular wheat cultivars of 1990s like CPAN3004, PBW175, PBW343, PBW373, UP2338, UP2382, UP2418 and UP2425 carrying 1BL/1RS translocation (Kumar et al. 2003) influenced the pathogen population dynamics leading to the breakdown of Yr9 resistance. This resulted in severe epidemics in Asia (Hodson 2011), and stripe rust began to emerge as a serious threat to wheat production in the region. In India, the first Yr9 virulence was detected in 1996 (pathotype 46S119) from the Gurdaspur area of Punjab and subsequently Yr9 + Yr27 virulence (pathotype 78S84) in 2001 (Prashar et al. 2007). The host differential line Tc*6/Lr26 (Yr9) became susceptible to these pathotypes, whereas cv. Riebesel 47/51 (Yr2, Yr9, +) remained resistant because it carried two or more additional stripe rust resistance genes. Stubbs and Yang (1988) found that cv. Riebesel 47/51 had Yr9 and at least one additional gene for resistance. Luthra et al. (1989) reported that cv. Riebesel 47/51 had four genes for resistance to Indian races. During 2013–14 cropping season, three new Yr9-virulent pathotypes 110S119, 110S84 and 238S119 possessing unique combined virulence for cvs Strubes Dickkopf (Yr2, Yr3a, Yr4a) and Suwon92xOmar (YrSU), were detected. Pathotype 238S119 had additional virulence for cv. Riebesel 47/51. Consequently, many resistant wheat cultivars became susceptible to stripe rust. Combined virulence for cvs Strubes Dickkopf and Suwon92xOmar have also been reported from North America (CDL2/97E96, CDL7/96E129, CDL24/100E132, CDL29/112E128, CDL31/98E139), Europe (106E139, 232E233), Syria (230E150, 230E134) and Lebanon (166E150) (de Vallavielle-Pope and Line 1990; Yahyaoui et al. 2002). Riebesel 47/51 and its derivative cultivars like Clement (Yr9, YrCle) and Lovrin 10 (Yr9), have been used as differentials to distinguish Pst races. Riebesel 47/51 has become susceptible to Pst pathotype/s in France, Clement in the USA, Canada, Ecuador and Lovrin 10 in China (de Vallavielle-Pope and Line 1990; Chen et al. 2002; Su et al. 2003; Wan et al. 2004; Ochoa et al. 2007).
There has been a rapid evolution of stripe rust pathogen in the North-Western Plains and Northern Hills Zones of India as new pathotypes have emerged almost every 5–6 years (Prashar et al. 2007; Gangwar et al. 2017). It was observed that with the emergence of new pathotypes, old ones were replaced gradually due to cultivation of new wheat cultivars which guided shifts in pathogen’s virulence patterns. For instance, wheat cultivar PBW 343 a cv. Attila sibling (ND/VG9144//KAL/BB/3/YACO/4/ VEE#5) was released in 1995 and, by 2002–03, it occupied more than 90% of the wheat-growing area of Punjab, and a total of seven million ha across the Indo-Gangetic Plains (Singh et al. 2017). The new pathotype 78S84, detected in 2001 showing virulence to cv. PBW 343, became frequent because of the large acreage under this cultivar. Since Berberis spp. do not play any role in the sexual cycle and perpetuation of the stripe rust fungus in India (Mehta 1940), we presume that mutation and adaptation to popular local cultivars are the most important mechanisms for the evolution and selection of new virulences (Gangwar et al. 2016). Pathotype 46S119 evolved independently from the known pathotype 46S103 by single step forward mutation (Prashar et al. 2007). It appears that a genetic change occurred recently in pathotype 46S119 resulting in additional virulence to cvs Suwon92xOmar and Riebesel 47/51. Similarly, pathotype 78S84 gained additional virulence to cv. Strubes Dickkopf. As a consequence of these changes, three new virulences evolved with combined virulence to cvs Strubes Dickkopf and Suwon92xOmar. Pathotypes/races with increased virulence factors are at an advantage in the pathogen population over those with fewer virulence factors because they are able to infect more wheat cultivars (Chen et al. 2010). This explains the breakdown of resistance in some wheat cultivars that previously were resistant to Pst pathotypes. Mutation is the basic cause of the occurrence of new virulences in countries where the stripe rust pathogen multiplies clonally. The dominance of such virulences will eventually depend upon their parasitic fitness and the opportunities to be selected through large-scale cultivation of cultivars with matching resistance (Chen et al. 2009).
Internal transcribed spacer (ITS) markers are informative and good phylogenetic tools to elucidate relationships among different variants of a pathogen. Therefore, we used universal primers (ITS1 and ITS4) for PCR amplification of rDNA containing the internal transcribed spacer, ITS1, 5·8S gene and ITS2 regions for probing into evolutionary pattern of new pathotypes of Puccinia striiformis f. sp. tritici. In drawing phylogenetic trees ITS 1 and ITS 2 can be used for distantly-related species whereas the 5.8S gene can be used for closely related species (Potkar and Jadhav 2015). In the present study, the new cv. Riebesel 47/51-virulent pathotype 238S119 showed close ITS sequences similarity with pathotype 46S119, which suggested the possibility of its evolution from the latter. Pathotypes 110S119 and 110S84 did not show such closeness. Hence they may have evolved independently.
Information regarding virulence of new Pst pathotypes is important for resistance breeding programmes toward effective stripe rust management (Sharma-Poudyal et al. 2013). Genetic resistance is a most effective, economic, and environmentally safe means of keeping the wheat stripe rust below the threshold level (Gangwar et al. 2017). Though triazole fungicides are effective in reducing losses due to wheat rust diseases, our priority is to minimize the use of chemicals in crop production from economical, ecological and health perspectives. For this reason, genetic resistance is the only pragmatic option for managing wheat rusts. Genetic engineering approaches are not cost-effective but the social acceptability of the products is low. In the present study, we identified 11 advanced lines showing resistance to new as well as the earlier reported Yr9-virulent pathotypes. These lines can be deployed for cultivation directly or used in stripe rust resistance breeding programmes.
References
Bhardwaj SC, Gangwar OP, Singh SB, Saharan MS, Sharma S (2012) Rust situation and pathotypes of Puccinia species in Leh Ladakh in relation to recurrence of wheat rusts in India. Indian Phytopathol 65(3):230–232
Bhardwaj SC, Prasad P, Gangwar OP, Khan H, Kumar S (2016) Wheat rust research-then and now. Indian J Agric Sci 86(10):123–144
Chen XM, Penman L (2005) Stripe rust epidemic and races of Puccinia striiformis in the United States in 2004. Phytopathology 95:S19
Chen XM, Moore M, Milus EA, Long DL, Line RF, Marshall D, Jackson L (2002) Wheat stripe rust epidemics and races of Puccinia striiformis f. sp. tritici in the United States in 2000. Plant Dis 86:39–46
Chen WQ, Wu LR, Liu TG, Xu SC, Jin SL, Peng YL, Wang BT (2009) Race dynamics, diversity, and virulence evolution in Puccinia striiformis f. sp. tritici, the causal agent of wheat stripe rust in China from 2003 to 2007. Plant Dis 93:1093–1101
Chen XM, Penman L, Wan A, Cheng P (2010) Virulence races of Puccinia striiformis f. sp. tritici in 2006 and 2007 and development of wheat stripe rust and distributions, dynamics, and evolutionary relationships of races from 2000 to 2007 in the United States. Can J Plant Pathol 32(3):315–333
Chen W, Wellings C, Chen X, Kang Z, Liu T (2014) Wheat stripe (yellow) rust caused by Puccinia striiformis f. sp. tritici. Mol Plant Pathol 15(5):433–446
Cooke DEL, Duncan JM (1997) Phylogenetic analysis of Phytophthora species based on ITS1 and ITS2 sequences of the ribosomal RNA gene repeat. Mycol Res 101:667–677
de Vallavielle-Pope C, Line RF (1990) Virulence of north American and European races of Puccinia striiformis on north American, world and European differential wheat cultivar. Plant Dis 74(10):739–743
Gangwar OP, Kumar S, Prasad P, Bhardwaj SC, Khan H, Verma H (2016) Virulence pattern and emergence of new pathotypes in Puccinia striiformis f. sp. tritici during 2011-15 in India. Indian Phytopathol 69(4s):178–185
Gangwar OP, Bhardwaj SC, Kumar S, Prasad P, Khan H, Savadi S (2017) Overcoming stripe rust of wheat: a threat to food security. In: Singh DP (ed) Management of wheat and barley diseases. Apple Academic Press, USA, pp 115–132
Hodson DP (2011) Shifting boundaries: challenges for rust monitoring. Euphytica 170:93–104
Kiran K, Rawal HC, Dubey H, Jaswal R, Bhardwaj SC, Prasad P, Pal D, Devanna BN, Sharma TR (2017) Dissection of genomic features and variations of three pathotypes of Puccinia striiformis through whole genome sequencing. Sci Rep 7:42419
Kumar S, Kumar N, Balyan HS, Gupta PK (2003) 1BL.1RS translocation in some Indian bread wheat genotypes and strategies for its use in future wheat breeding. Caryologia 56(1):23–30
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Larkin MA, Blackshields G, Brown NP, Chenna R et al (2007) CLUSTALW and CLUSTALX version 2. Bioinformatics 23:2947–2948
Lee SB, Taylor JW (1992) Phylogeny of five fungus-like protoctistan Phytophthora species, inferred from the internal transcribed spacer of ribosomal DNA. Mol Biol Evol 9:636–653
Luthra JK, Prabhu KV, Verma RPS (1989) Inheritance of resistance to stripe rust (Puccinia striiformis) in some yellow rust resistance tester of wheat (Triticum aestivum). Indian J Agric Sci 59:617–621
McIntosh RA, Wellings CR, Park RF (1995) Wheat rusts: an atlas of resistance genes. CSIRO Publishing, Melbourne, p 199
Mehta KC (1940) Further studies on Cereal Rusts in India (Vol I). Scientific Monograph 14, Imperial Council Agricultural Research, New Delhi
Nagarajan S, Joshi LM (1975) A historical account of wheat rust epidemics in India and their significance. Cereal Rusts Bull 3:29–33
Nagarajan S, Bahadur P, Nayar SK (1984) Occurrence of new virulence 47S102 of Puccinia striiformis west., In India during crop year 1982. Cereal Rusts Bulletin 27: 28–31
Nagarajan S, Nayar SK, Bahadur P (1985) The proposed system of virulence analysis. III. Puccinina striiformis west. Kavaka 13(1):33–36
Nayar SK, Prashar M, Bhardwaj SC (1997) Manual of current techniquesin wheat rust. Research bulletin 2, Regional Station, Directorate of Wheat Research, Flowerdale Shimla, Himachal Pradesh, India. pp 1–32
Ochoa JB, Danial DL, Paucar B (2007) Virulence of wheat yellow rust races and resistance genes of wheat cultivars in Ecuador. Euphytica 153:287–293
Potkar VR, Jadhav PS (2015) Phylogenetic analysis and predicted secondary structure of 5.8s gene in Puccinia graminis f. sp. tritici. Int J Inst Pharm Life Sci 5(2):104–111
Prashar M, Bhardwaj SC, Jain SK, Datta D (2007) Pathotypic evolution in Puccinia striiformis in India during 1995-2004. Aust J Agric Res 58:602–604
Prescott JM, Burnett PA, Saari EE, Ransom J, Bowman J, Milliano W, Singh RP, Bekele G (1986) Wheat Diseases and Pests: A Guide for Field Identification. Mexico, D. F.: International Maize and Wheat Improvement Center (CIMMYT)
Sharma-Poudyal D, Chen XM, Wan AM, Zhan GM, Kang ZS, Cao SQ, Jin SL, Morgounov A, Akin B, Mert Z, Shah SJA, Bux H, Ashraf M, Sharma RC, Madariaga R, Puri KD, Wellings C, Xi KQ, Wanyera R, Manninger K, Ganzalez MI, Koyda M, Sanin S, Patzek LJ (2013) Virulence characterization of international collections of the wheat stripe rust pathogen, Puccinia striiformis f. sp. tritici. Plant Dis 97:379–386
Singh RP, Srivastava P, Sharma A, Bains NS (2017) Bread wheat cultivar PBW 343 carries residual additive resistance against virulent stripe rust pathotype. J Crop Improv 31(2):183–191
Stubbs RW, Yang HA (1988) Pathogenicity of Puccinia striiformis for wheat cultivars with resistance derived from rye. Proc Eur Meditter Cereal Rusts conf 7:110–112
Su H, Conner RL, Graf RJ, Kuzyk AD (2003) Virulence of Puccinia striiformis f. sp. tritici, cause of stripe rust on wheat, in western Canada from 1984 to 2002. Can J Plant Pathol 25:312–319
Tamura K (1992) Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol Biol Evol 9:678–687
Wan A, Zhao Z, Chen X, He Z, Jin S, Jia Q, Yao G, Yang J, Wang B, Li G, Bi Y, Yuan Z (2004) Wheat stripe rust epidemic and virulence of Puccinia striiformis f. sp. tritici in China in 2002. Plant Dis 88:896–904
White TJ, Bruns TD, Lee SB, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Yahyaoui AH, Hakim MS, El Niami M and Rbeiz N (2002) Evolution of physiologic races and virulence of Puccinia striiformis on wheat in Syria and Lebanon. Plant Dis 86(5):499–504
Yahyaoui AH, Hakim MS, El Niami M and Rbeiz N (2002) Evolution of physiologic races and virulence of Puccinia striiformison wheat in Syria and Lebanon. Plant Dis 86(5):499–504
Zeller FJ, Sears ER, Sears LMS (1973) 1B/1R wheat-rye chromosome substitutions and translocations. In: Sears ER, LMS S (eds) Proceedings of the fourth international wheat genetics symposium. University of Missouri, Columbia, pp 209–221
Acknowledgements
We thank to the Director, ICAR-Indian Institute of Wheat and Barley Research, Karnal, Haryana, India, for providing liberal support to undertake the survey work. We are very grateful to Dr. A.N. Mishra, Emeritus Scientist, ICAR- IARI, Regional Station, Indore, India, for critical appraisal of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 35 kb)
Rights and permissions
About this article
Cite this article
Gangwar, O.P., Kumar, S., Bhardwaj, S.C. et al. Characterization of three new Yr9-virulences and identification of sources of resistance among recently developed Indian bread wheat germplasm. J Plant Pathol 101, 955–963 (2019). https://doi.org/10.1007/s42161-019-00302-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42161-019-00302-w