Abstract
The ratio of human to bacterial cells in the human body (microbiota) is around 1:1. As a result of co-evolution of the host mucosal immune system and the microbiota, both have developed multiple mechanisms to maintain homeostasis. However, dissociations between the composition of the gut microbiota and the human host may play a crucial role in the development of type 2 diabetes. Metformin, the most frequently administered medication to treat patients with type 2 diabetes, has only recently been suggested to alter gut microbiota composition through the increase in mucin-degrading Akkermansia muciniphila, as well as several SCFA-producing (short-chain fatty acid) microbiota. The gut microbiota of participants on metformin has exerted alterations in gut metabolomics with increased ability to produce butyrate and propionate, substances involved in glucose homeostasis. Thus, metformin appears to affect the microbiome, and an individual’s metformin tolerance or intolerance may be influenced by their microbiome. In this review, we will focus on the effects of metformin in gut microbiota among patients with T2DM.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The number of microorganisms in the human intestine is approximately 1 × 1014, while the number of eukaryotic cells in the human body is around 1 × 1013. The number of microbes inhabiting the intestines has been estimated to be 1014, which encompasses approximately 10 times more bacterial cells than the number of human cells and over 100 times the amount of genomic content (microbiome) as compared to the human genome [1, 2]. Meanwhile, a more recent estimation has demonstrated that the ratio of human to bacterial cells in the human body (microbiota) is, in fact, around 1:1 [3]. As a result of co-evolution of the host mucosal immune system and the microbiota, both have developed multiple mechanisms to maintain homeostasis [3]. However, dissociations between the composition of the gut microbiota and the human host may play a crucial role in the development of type 2 diabetes (T2DM) [4,5,6,7]. Large metagenome-wide studies in China and Europe have documented gut microbial dysbiosis associated with obesity and T2DM. The common finding of these studies was a reduction in butyrate-producing microbes, together with an increase in opportunistic pathogens [4,5,6,7,8]. Data from these studies and the Danish MetaHIT project were analyzed after controlling for metformin administration [4,5,6,7,8,9]. The conclusion was that the noted reduction in butyrate-producing bacteria could imply gut microbiome shifts among patients with T2DM [4,5,6,7,8,9]. On the other hand, while recent data suggest that metformin alters gut microbiota, the classic mechanisms of action, such as inhibition of hepatic gluconeogenesis, still retain their importance in the treatment of patients with T2DM [4,5,6,7,8,9].
Metformin and metabolomics
Metformin, the most frequently administered medication to treat patients with T2DM, has only recently been suggested to alter gut microbiota composition through the increase in mucin-degrading Akkermansia muciniphila, as well as several SCFA-producing (short-chain fatty acid) microbiota [10]. A meta-analysis of 199 individuals with T2DM and 544 control participants without diabetes confirmed that metformin significantly altered gut microbiota composition: metformin patients with T2DM could be identified based on compositional changes in their gut microbiota [9].
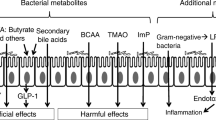
Metabolomics, i.e., the scientific study of chemical processes involving metabolites—the small molecule intermediates and products of metabolism—are likely to shed much light on this subject [11]. Specific microbiome profiles are related to increased glucose intolerance [12]. Changes in gut microbiota may influence the gut metabolome, thus affecting the intestinal output of butyrate and acetate [12, 13]. These are major gut-derived metabolites which are implicated in the host’s insulin resistance and, thus, glycemic control. It has been suggested that an enhancement in bacteria producing the short-chain fatty acids butyrate and propionate may improve glycemia (Fig. 1) [14]. Butyrate and propionate increase intestinal gluconeogenesis. In rodents, increased intestinal gluconeogenesis results in a reduction in hepatic gluconeogenesis, appetite, and weight, leading to improved glucose homeostasis. In particular, the gut microbiota of participants on metformin exerted increased ability to produce butyrate and propionate, while in the untreated individuals, there was enhancement in microbial genes implicated in the degradation of glycine and tryptophan as well as decreases in genes involved in threonine and arginine degradation. This is of particular interest since glycine has been associated with insulin sensitivity, and glycine supplementation has been reported to improve insulin sensitivity in T2DM [12, 13].
Metformin and intestinal absorption
There is a growing body of evidence in favor of a key role of metformin in the lower gut [15,16,17,18,19,20,21,22,23]. It is noteworthy that metformin levels in the gut are 100- to 300-fold higher than those in serum, making the gut the primary reservoir for metformin in humans [15,16,17,18,19,20,21,22,23]. Unlike oral administration, intravenous administration of metformin in humans does not improve glycemia [22]. Rates of systemic absorption and intestinal transit time affect the concentrations of metformin to which the microbiota are exposed: oral bioavailability is about 30–60%, and approximately 50% of an ingested dose is found in the feces, although both these factors exhibit significant person-to-person variability. Thus, the degree to which the intestinal flora is altered by the drug is affected by host factors [15,16,17,18,19,20,21,22,23].
In particular, the oral absorption and hepatic uptake of metformin is performed by organic cation transporters (OCTs), mainly OCT1 and OCT3, whereas renal excretion of metformin is mediated by OCT2 [24,25,26]. A study performed in rodents has demonstrated that metformin could be a superior substrate for renal OCT2 rather than for hepatic OCT1 [27]. Besides, OCT2 accounts for gender differences in renal organic cation transporting activity [28,29,30]. Furthermore, OCT1 is also highly polymorphic: 34 OCT1 polymorphisms have been identified among 10 ethnic groups. Met408Val (rs628031) variant has been the most frequently explored as regards metformin effects. Although some alleles have been linked to negative effects on metformin, others have shown positive effects [31]. These polymorphisms provide a plausible explanation for the interindividual variations in metformin response [31]. However, is there any relationship between polymorphisms as concerns OCTs, metformin, and microbiota? Only recently, a study demonstrated that differences in abundance of Bifidobacterium could be explained by opioids acting as organic cation transporter 1 (OCT1) inhibitors [32]. This study was the first to document that abundance of Bifidobacterium and Prevotella was influenced by interactions between T2DM and the use of metformin and opioid (acting as OCT inhibitors) [32]. Therefore, the issue of metformin, OCT, and gut microbiota remains to be further investigated in the near future. The pharmacological actions of metformin include changes in bile acid recirculation and gut microbiota, resulting in enhanced levels of glucagon-like polypeptide-1 (GLP-1). These observations have been confirmed by clinical studies testing the response to delayed-release metformin (MetDR) in healthy participants and in patients with T2DM [33,34,35]. The delayed release of metformin relies on pH-dependent dissolution of the enteric coat of the tablet; this mainly occurs at pH 6.5 in the distal intestine, where there is abundancy of L cells, which accounts for the gut-based mechanisms of metformin. Studies have documented that MetDR has a glucose-lowering effect, which is comparable to that of extended release metformin, but with a 40% increase in its potential, thus allowing for lower doses of MetDR to be used [33,34,35]. Moreover, two randomized trials have shown that MetDR resulted in glucose-lowering effects comparable to immediate-release metformin, with lower systemic exposure [33,34,35]. Nevertheless, once-daily dosing with MetDR 1500 mg results in improved glucose-lowering effect, with immediate-release metformin 1000 mg twice daily and better gastrointestinal tolerance [33,34,35,36]. Whether MetDR affects the gut microbiota in a manner similar to that of immediate-release metformin remains unknown and needs to be elucidated [33,34,35,36].
Metformin and its adverse gastrointestinal effects
The work by Forslund et al. postulates that microbiota-mediated mechanisms lie behind both the therapeutic and adverse effects of metformin [9]. Alterations in the microbiome by metformin may account for its gastrointestinal intolerance. Burton et al. carried out a study using a gastrointestinal microbiome modulator (GIMM) or placebo together with metformin in metformin-intolerant patients with T2DM [33]. They found that treatment with metformin in conjunction with GIMM resulted in lower fasting glucose levels, suggesting better tolerance of metformin therapy for longer or at a higher dose. Thus, metformin appears to affect the microbiome, and an individual’s metformin tolerance or intolerance may be influenced by their microbiome. Furthermore, increases in the genus Escherichia in metformin-treated individuals have been functionally associated with gas metabolism indicating involvement of the genus Escherichia in the adverse gastrointestinal effects of metformin [37].
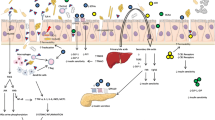
Metformin’s main adverse side effect, i.e., lactic acidosis, is suggested as being due to the accumulation of lactate via the inhibition of hepatic glucose production from lactate molecules. Moderate to severe renal impairment as well as drugs inhibiting the metformin transporters (OCTs) could lead to the development of lactic acidosis, with potentially fatal results, due to an increase in its plasma concentrations [38, 39]. Recently, the 808G>T variance in the SLC22A2 gene can affect the plasma lactate level and the incidence of hyperlactacidemia in T2DM patients on metformin treatment [40] (Fig. 2). In addition, the effect of metformin on gut microbiota among patients with GFR < 60 ml/min/1.73 m2 remains unknown, as studies dealing with this particular issue are lacking.
Conclusions
It must be recognized that metformin may have an impact on systemic carbohydrate metabolism via gastroenteric mechanisms that do not directly involve gut microbiota. These include changes in bile acid physiology, enteric hormones, and effects on duodenal AMPK signaling, which contribute to suppression of hepatic gluconeogenesis [34,35,36]. While recent data suggest that metformin alters gut microbiota, the classic mechanisms of action, such as inhibition of hepatic gluconeogenesis, still retain their importance in the treatment of patients with T2DM. Further research is thus mandatory to evaluate the effects of metformin, which may be attributed to changes in gut microbiota. It is anticipated that these effects will vary considerably among metformin-treated patients.
References
Backhed F (2005) Host-bacterial mutualism in the human intestine. Science 307:1915–1920
Gill SR, Pop M, DeBoy RT et al (2006) Metagenomic analysis of the human distal gut microbiome. Science 312:1355–1359
Sender R, Fuchs S, Milo R (2016) Revised estimates for the number of human and bacteria cells in the body. PLoS Biol 14(8):e1002533
Qin J, Li Y, Cai Z et al (2012) A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490:55–60
Karlsson FH, Tremaroli V, Nookaew I et al (2013) Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498:99–103
Larsen N, Vogensen FK, van den Berg FWJ et al (2010) Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One 5:e9085
Zhang X, Shen D, Fang Z et al (2013) Human gut microbiota changes reveal the progression of glucose intolerance. PLoS One 8:e71108
Tilg H, Moschen AR (2014) Microbiota and diabetes: an evolving relationship. Gut 63:1513–1521
Forslund K, Hildebrand F, Nielsen T et al (2015) Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528:262–266
de la Cuesta-Zuluaga J, Mueller NT, Vanessa Corrales-Agudelo V et al (2017) Metformin is associated with higher relative abundance of mucin-degrading Akkermansia muciniphila and several short-chain fatty acid-producing microbiota in the gut. Diabetes Care 40:54–62
Bennett D (2005) Growing pains for metabolomics. Scientist 19(8):25–28
Pedersen HK, Gudmundsdottir V, Nielsen HB et al (2016) Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 535:376–381
Perry RJ, Peng L, Barry NA et al (2016) Acetate mediates a microbiome-brain-beta-cell axis to promote metabolic syndrome. Nature 534:213–217
Lee H, Ko G (2014) Effect of metformin on metabolic improvement and gut microbiota. Appl Environ Microbiol 80:5935–5943
Shin NR, Lee JC, Lee HY et al (2014) An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 63:727–735
Napolitano A, Miller S, Nicholls AW et al (2014) Novel gut-based pharmacology of metformin in patients with type 2 diabetes mellitus. PLoS One 9:e100778
Gall WE, Beebe K, Lawton KA et al (2010) α-Hydroxybutyrate is an early biomarker of insulin resistance and glucose intolerance in a nondiabetic population. PLoS One 5:e10883
Bonora E, Cigolini M, Bosello O et al (1984) Lack of effect of intravenous metformin on plasma concentrations of glucose, insulin, C-peptide, glucagon and growth hormone in non-diabetic subjects. Curr Med Res Opin 9:47–51
Bailey CJ, Wilcock C, Scarpello JH (2008) Metformin and the intestine. Diabetologia 51:1552–1553
Sekar S, Chandrasekaran A, Rao U, Sastry TP (2011) Comparison of some of the physicochemical characteristics of type 2 diabetic and normal human bones: a sample study. J Diabetes Complicat 25:187–192
DeFronzo RA, Buse JB, Kim T et al (2016) Once-daily delayed release metformin lowers plasma glucose and enhances fasting and postprandial GLP-1 and PYY: results from two randomised trials. Diabetologia 59:1645–1654
Buse JB, DeFronzo RA, Rosenstock J et al (2016) The primary glucose-lowering effect of metformin resides in the gut, not the circulation: results from short-term pharmacokinetic and 12-week dose-ranging studies. Diabetes Care 39:198–205
Wu H, Esteve E, Tremaroli V et al (2017) Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med. https://doi.org/10.1038/nm.4345
Gong L, Goswami S, Giacomini KM, Altman RB, Klein TE (2012) Metformin pathways: pharmacokinetic and pharmacodynamics. Pharmacogenet Genomics 22(11):820–827
Dresser MJ, Xiao G, Leabman MK, Gray AT, Giacomini KM (2002) Interactions of N-tetraalkylammonium compounds and biguanides with a human renal organic cation tranporter (hOCT2). Pharm Res 19:1244–1247
Kimura N, Masuda S, Tanihara Y et al (2005) Metformin is a superior substrate for renal organic cation tranporter OCT2 rather than hepatic OCT1. Drug Metab Pharmacokinet 20:379–386
Urakami Y, Nakamura N, Takahashi K et al (1999) Gender differences in expression of organic cation transporter OCT2 in rat kidney. FEBS Lett 461:339–342
Urakami Y, Okuda M, Saito H, Inui K (2000) Hormonal regulation of organic cation transporter OCT2 expression in rat kidney. FEBS Lett 473:173–176
Asaka J, Terada T, Okuda M, Katsura T, Inui K (2006) Androgen receptor is responsible for rat organic cation transporter 2 gene regulation but not for rOCT1 and rOCT3. Pharm Res 23:697–704
Leabman MK, Giacomini KM (2003) Estimating the contribution of genes and environment to variation in renal drug clearance. Pharmacogenetics 13:581–584
Mofo Mato EP, Guewo-Fokeng M, Essop MF, Oroma Owira PM (2018) Genetic polymorphisms of organic cation transporter 1 (OCT1) and responses to metformin therapy in individuals with type 2 diabetes: a systematic review. Medicine 97:27e11349
Barengolts E, Green SJ, Eisenberg Y et al (2018) Gut microbiota varies by opioid use, circulating leptin and oxytocin in African American men with diabetes and high burden of chronic disease. PLoS One 13(3):e0194171
Burton JH, Johnson M, Johnson J, Hsia DS, Greenway FL, Heiman ML (2015) Addition of a gastrointestinal microbiome modulator to metformin improves metformin tolerance and fasting glucose levels. J Diabetes Sci Technol 9:808–814
Brunkwall L, Orho-Melander M (2017) The gut microbiome as a target for prevention and treatment of hyperglycaemia in type 2 diabetes: from current human evidence to future possibilities. Diabetologia 60:943–951
McCreight LJ, Bailey CJ, Pearson ER (2016) Metformin and the gastrointestinal tract. Diabetologia 59:426–435
Henry RR, Frias JP, Walsh B et al (2018) Improved glycemic control with minimal systemic metformin exposure: effects of metformin delayed-release (metformin DR) targeting the lower bowel over 16 weeks in a randomized trial in subjects with type 2 diabetes. PLoS One 13(9):e0203946
Duca FA, Cote CD, Rasmussen BA et al (2015) Metformin activates a duodenal AMPK-dependent pathway to lower hepatic glucose production in rats. Nat Med 21:506–511
Duong JK, Furlong TJ, Roberts DM et al (2013) The role of metformin in metformin-associated lactic acidosis (MALA): case series and formulation of a model of pathogenesis. Drug Saf 36(9):733–746
Maideen NMP, Jumale A, Balasubramaniam R (2017) Drug interactions of metformin involving drug transporter proteins. Adv Pharm Bull 7(4):501–505
Li Q, Liu F, Zheng TS, Tang JL, Lu HJ, Jia WP (2010) SLC22A2 gene 808 G/T variant is related to plasma lactate concentration in Chinese type 2 diabetics treated with metformin. Acta Pharmacol Sin 31:184–190
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Human and animal rights and informed consent
This article does not contain any studies with human participants performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Vallianou, N.G., Stratigou, T. & Tsagarakis, S. Metformin and gut microbiota: their interactions and their impact on diabetes. Hormones 18, 141–144 (2019). https://doi.org/10.1007/s42000-019-00093-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42000-019-00093-w