Abstract
Over the past few decades, the enzyme-mimicking activity of metal–organic frameworks (MOFs) accompanied with structural characteristics has aroused much attention. However, pure MOFs have low affinity with DNA. Here, iron-based MOFs with acidized carbon nanotubes (CNTs) via a simple hydrothermal process have been synthesized, named as MIL-88(NH2)-Fe@CNTs. CNTs can enhance the affinity between MOF and DNA, achieving flexible regulation of their catalytic activity benefiting from the strong π − π stacking between CNTs and DNA. Meanwhile, in comparison with conventional iron-based MOFs, the addition of CNTs, which contributes to the acceleration of electron transfer, endowing as-prepared nanocomposites remarkably enhanced peroxidase-like activity to achieve an ultrasensitive detection of H2O2 with the LOD of 17.64 μg/L. Notably, the as-prepared nanocomposites with adsorbed DNA displayed excellent affinity towards both TMB (3, 3′, 5, 5′-tetramethylbenzidine) substrates and H2O2 as well as high catalytic velocity. On the basis of their switchable peroxidase-like activity regulated by different length or sequence of ssDNA, it is believed that our-prepared MOF-based nanomaterials would be promising for fabricating versatile and sensitive label-free colorimetric assays for diverse targets.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Nanozymes, a new kind of artificial enzyme based on nanomaterials, which possess low cost, high stability and adjustable catalytic activity compared to natural enzyme, has always been an interesting topic [1,2,3,4,5]. To date, a wealth of nanomaterials including carbon-based nanomaterials, metal oxides, nanoparticles, and MOFs have been reported to possess unique enzyme-like catalytic properties and widely explored for various purposes, including environment monitoring, bio-sensing, disease diagnosing and therapeutics [6,7,8,9]. Especially, researchers have put great efforts on mimicking oxidase or peroxidase and successfully explored their applications in sensing field. Recently, MOFs, emerging as a subclass of porous coordination polymers, have shown the high potential of in storage, chemical catalysts and biosensor [10,11,12]. Particularly, benefiting from their unique structure of providing a large number of biomimetic active sites accessible to substrates, a series of MOFs have been developed as different enzyme mimics and successfully applied in sensing field [13, 14]. In addition, some iron-based MOFs have also been employed as peroxidase mimics and developed for colorimetric detection of H2O2, glucose and some other reducing agents [15,16,17]. However, most of current works are difficult to meet the requirements of high sensitivity and high selectivity at the same time, which may hinder the potential applications of MOFs-based enzyme mimics. Therefore, the construction of a sensitive and specific sensing platform for detecting various targets on the basis of MOFs-based peroxidase mimics still needs discussion.
Notably, aptamers are a kind of functional DNA or RNA structures commonly used as recognition elements, which are able to recognize specific targets [18]. Inspired by this, the specific detection of diverse targets was realized by using matched aptamers/targets integrating with switchable enzyme-like activities [19]. Nanozymes with DNA are considered as an appropriate way to build sensing platforms with recognition elements [20]. Through surface functionalization, MOFs may have a better affinity to DNA [21, 22]. Generally, it was found that carbon-based nanomaterials such as graphene, CNTs and carbon dots exhibit high affinity to biomolecules (e.g., nucleic acids, antibodies and protein) [23], which provides a new idea to improve the interaction between MOFs and DNA. Among them, carbon nanotubes (CNTs) have tubular morphology with the merits of large surface area, excellent conductivity and great stability, which attract extensively attention in various fields [24, 25]. More importantly, the peroxidase-like CNTs can be regulated by DNA and applied for exosome colorimetric detection [26]. Besides, acidified CNTs with better water-solubility owing to their rich carboxyl and hydroxyl groups [27], which holds great potential for expanding application range of their composite materials. In addition, the surface of CNTs was easily functionalized by a large number of oxygen functional groups (e.g., –COOH, –OH, etc.) [28, 29]. The modification of MOFs with CNTs is regarded as an effective way to acquire dramatically improved catalytic performance. For example, Peng reveals that Low concentration MWCNTs-COOH exhibited good catalytic ability to TCS (triclosan) oxidation [30]. Very recently, MOFs/CNTs nanocomposites have been explored as peroxidase mimics for visual detection. The utilization of CNTs can improve peroxidase mimics catalysis process [31]. Despite the above process, the peroxidase-like of CNTs modified with MOFs may be regulated through altering DNA, which may further broaden the possible application range of nanozymes with high selectivity.
Herein, we have synthesized iron-based MOFs modified with CNTs via a hydrothermal route. CNTs enhance the affinity of MOFs to DNA. The flexible regulation of peroxidase-like activity of CNTs modified MOFs achieved through altering DNA sequence, length and concentration. The corresponding regulated mechanism was discussed. In addition, CNTs improved peroxidase-like of MOFs, and the composites achieve a simple detection of H2O2. Also, the change of bandgap before and after modification have been discussed. Furthermore, MOF/CNTs modified with DNA are anticipated to employ the as-prepared nanocomposites to develop label-free, sensitive and highly specific colorimetric platforms.
2 Experimental
2.1 Materials and Chemicals
DNA oligomers were provided by Sangon Biotech Co., Ltd (Shanghai, China). TMB was obtained from Aladdin Reagent Co., Ltd (Shanghai, China). Carbon nanotubes with diameter of 10-20 nm (CNTs purity > 97%, length > 5 nm) were provided by Shenzhen Nanotech Pore Co., Ltd. Sulfuric acid, nitric acid and sodium hydroxide were purchased from Tianjin Fuyu Fine Chemical Co., Ltd. The other chemicals [ferric chloride hexahydrate (FeCl3·6H2O), 2-aminoterephthalic acid, acetic acid (CH3COOH), acetic acid, sodium salt (CH3COONa), DMF, methanol] were obtained from Sinopharm Chemical Reagent Co., Ltd (Shanghai, China) and used without further purification. All solutions were prepared with ultrapure water from Millipore water purification system (Laikie Instrument Co., Ltd., Shanghai, China).
2.2 Instruments
The morphologies of the samples were acquired by using a field-emission scanning electron microscopy (SEM, HitachiS-4800). X-ray diffraction (XRD) was performed on a Rigaku SmartLab 9 with Cu kα radiation at 45 kV and 200 mA. The chemical structures of as-prepared samples were obtained through Fourier Transform Infrared (FT-IR, iN10). Jasco V-550 spectrometer was employed for ultraviolet–visible (UV–vis) absorption spectra. Zeta Potential was obtained through Laser Particle Size and Zeta Potential Analyzer (zs 90). UV–vis spectrophotometer (UV-2450) was employed for diffuse reflectance spectra. Transmission electron microscopy (TEM) images were gained from high-magnification TEM (FEI Tecnai G2 F30 S-Twin). Fluorescence intensity was gained from Fluorescence spectrophotometer (F-4500).
2.3 Preparation of CNTs
The preparation of CNTs was based on reported work [32]. 10 mL nitric acid and 30 mL sulfate acid were added into 1 g CNTs at 80 °C for 30 min. After natural cooling, CNTs was collected by filtration and washed by ultrapure water several times until pH of the filtrate was nearly neutral. Lastly, CNTs were dried in the vacuum freeze drier at 60 °C for 12 h.
2.4 Synthesis of MIL-88(NH2)-Fe@CNTs
The synthesis of MIL-88(NH2)-Fe@CNTs was followed by a previously reported method with some modification [33]. 2-Aminoterephthalic Acid (0.725 g) was dispersed in DMF (20 mL). The mixture and 4 M NaOH (0.8 mL) were injected dropwise for continuous stirring. Then, FeCl3·6H2O (1.081 g) and acidified CNTs (200 mg) were added. The mixture was stirred for 30 min. Subsequently, the above mixture was transferred into 50 mL of autoclave and hydrothermal reaction for 12 h at 100 °C. After reaction, the materials were collected by centrifugation and washed by DMF, methanol and ultrapure water for several times. The materials were dried in the vacuum freeze drier at 60 °C for 12 h.
2.5 DNA Adsorption and its Flexible Regulation of Peroxidase-like Catalytic Activity
For analyzing the effect of CNTs on the peroxidase-like catalytic properties of MIL-88(NH2)-Fe, TMB (0.18 mM) and H2O2 (0.18 mM) were mixed with DNA, MIL-88(NH2)-Fe and MIL-88(NH2)-Fe@CNTs in 0.20 M acetate buffer (pH 4.00), respectively. DNA with different bases (A22, T22, C22, or G22, 0.29 μM each), Poly-G DNA (2, 5, 10, 22, 30 mer, 8.824 μM) and Poly-G DNA (22 mer) with different concentration (1, 3, 5, 10, 15 μM) were incubated with MIL-88(NH2)-Fe@CNTs (0.03 mg/mL) in shaker at 37 °C for 2 h. Then, TMB (0.18 mM) and H2O2 (0.18 mM) were added into the mixture with acetate buffer (pH 4.00) at 37 °C. To investigate the influence of pH, MIL-88(NH2)-Fe@CNTs (0.03 mg/mL) and poly G22 (0.29 μM) were incubated in shaker at 37 °C for 2 h. TMB (0.18 mM) and H2O2 (0.18 mM) were mixed with above mixture in 0.20 M acetate buffer (pH 4, 5, 6, 7, 8), respectively. All of the reaction conditions of the above experiments were at 37 °C for 10 min. The UV–vis absorption spectra of the solution were recorded with a UV–vis spectrometer in absorbance-dependent mode.
For exploring the effect of CNTs on the affinity of MIL-88(NH2)-Fe to DNA, DNA (Pb-sub-NO) labeled by FAM was mixed with MIL-88(NH2)-Fe and MIL-88(NH2)-Fe@CNTs respectively in shaker at 37 °C for 2 h, after that, the supernatants of mixture were obtained by centrifugation and the fluorescence intensity of supernatant was measured.
H2O2detection: MIL-88(NH2)-Fe@CNTs (0.029 mg/L), TMB (0.18 mM) and H2O2 (0.18 mM) at different concentrations were mixed in 0.20 M acetate buffer. After reaction at 37 °C for 10 min, the UV–vis absorption spectra of the solution were recorded with a UV–vis spectrometer in absorbance-dependent mode.
3 Result and Discussion
3.1 Characterization
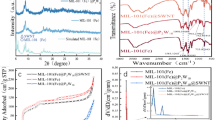
MIL-88(NH2)-Fe was obtained on the basis of previously reported method with a rods-like shape. From the SEM image in Fig. 1, our-prepared MIL-88(NH2)-Fe@CNTs still maintain the original morphology, evenly distributed, with the introduction of acidized CNTs. Crystalline structures of as-prepared samples were characterized by XRD. Several peaks of MIL-88(NH2)-Fe are agree with the reported works [34,35,36,37]. The similar diffraction peaks between MIL-88(NH2)-Fe and MIL-88(NH2)-Fe@CNTs in Fig. 2a reveals that the crystal structure of the prepared nanocomposites have no obvious change. To explore the effect of modification on the chemical structure especially the functional groups, FT-IR was performed. The peaks located at 769 cm−1 and 1258 cm−1 were clearly observed on two samples, which should attribute to C-H bending vibration of benzene ring and C–N, respectively. And the peaks corresponding to the asymmetric stretching of the carboxyl groups were also identified at 1383 cm−1 and 1575 cm−1. Note that the peaks of CNTs are invisible [38], maybe due to the small content of SWNTs. In MIL-88(NH2)-Fe@CNTs, the peaks show slight movements, indicating the effective hydrogen bonds and π–π interactions between MIL-88(NH2)-Fe and CNTs [31]. In this regard, the structure of CNT-modified MIL-88(NH2)-Fe remains unchanged. The difference between MIL-88(NH2)-Fe and MIL-88(NH2)-Fe@CNTs can been seen through TEM. As depicted in TEM image (Fig. 3), it was found that some tubular architecture (CNTs) interweave with the MIL-88(NH2)-Fe of rods-like shape. The above results reveal the successful synthesis of MIL-88(NH2)-Fe@CNTs.
3.2 Effect of CNTs on the Peroxidase-like Activity of MIL-88(NH2)-Fe with or without DNA
It was found that the peroxidase-like activity of MOFs could be promoted or inhibited by DNA through influencing the interaction between the enzyme mimics and substrates [39,40,41,42]. Hence, based on the typical TMB-H2O2 catalytic chromogenic reaction, the effect of CNTs on the catalytic activity of MIL-88(NH2)-Fe with or without DNA was explored. As shown in Fig. 4, the peroxidase-like activities of as-prepared samples were determined with TMB as the substrates in the presence of H2O2. The typical absorption band of oxidation state of TMB (oxTMB) at 652 nm was clearly observed in the system containing catalysts. Interestingly, the peroxidase-like activity of CNT-modified MIL-88(NH2)-Fe was superior to that of MIL-88(NH2)-Fe alone in the absence of DNA (curves c and d). Here, the Tauc plots were fitted in Fig. 5. The band gaps of MIL-88(NH2)-Fe and MIL-88(NH2)-Fe@CNTs were tested to be 1.25 eV and 0.33 eV. The narrower band gap promotes a faster electron transfer process, and thus helps the enhancement of catalytic activity. In other words, the modification of CNTs can effectively improve the catalytic activity of MIL-88(NH2)-Fe, which is consistent with reported works [31]. Therefore, based on the higher peroxidase-like activity of our-prepared nanocomposites, the ultrasensitive detection of H2O2 as an example was achieved. As shown in Fig. 6a, the absorbance intensity at 652 nm (typical absorption peak of oxTMB) increased with the increasing concentration of H2O2. And the detection limit (LOD) of H2O2 was calculated to 17.64 μg/L (3σ/k), representing the highly sensitive performance of the MIL-88(NH2)-Fe@CNTs-based colorimetric sensors.
Furthermore, it was confirmed that the CNT-modified nanocomposites further enhance the oxidation degree towards TMB substrates while the adsorption of DNA has brought the higher peroxidase-like catalytic activity (curves a and b in Fig. 4). Accordingly, it can be inferred that the desirable affinity of CNTs to DNA via π − π stacking strengthen the adsorption of DNA on the MIL-88(NH2)-Fe, which contributes to the TMB substrates binding and catalytic activity enhancement.
3.3 Possible Mechanism Analysis
As mentioned above, DNA adsorption is an important factor affecting catalytic activity. In the following experiments, ssDNA (single-stranded DNA) was adopt unless otherwise stated. First, the effect of different sequence of DNA on activity enhancement was explored. As shown in Fig. 7a, poly-C22 and poly-G22 effectively promoted the catalytic oxidation of TMB substrates. However, similar to condition of without DNA, both poly-A22 and poly-T22 have no effect on the catalytic activity, which may be contributed to the acidic environment in this reaction system. According to the pKa values, protonation of guanine and cytosine enabled the surface charges change, thus strengthening the interaction between the catalysts and DNA and the catalytic oxidation of TMB. G22 has regulation on both of MIL-88(NH2)-Fe and MIL-88(NH2)-Fe@CNTs, while MIL-88NH2)-Fe@CNTs with G22 has a better regulation (Figure S2). Moreover, the improved catalytic activity was also related to the length of DNA. One can see that the oxidation degree of TMB gradually enhances with the increasing DNA length in Fig. 7b, showing that the poly-G30 with more binding sites exhibited the strongest effect on the catalytic activity. In addition, the absorbance intensity of oxTMB did no longer increase when the concentration of DNA beyond 5 μM (Fig. 7c), which may be caused by the saturation of DNA on the surface of MIL-88(NH2)-Fe@CNTs.
Considering that pH value may play an important role in the catalytic reaction, the oxidation of TMB under different pH ranging from 4 to 8 was determined. From the Fig. 7d, it can be concluded that the adsorption of DNA on the prepared MIL-88(NH2)-Fe@CNTs depends on pH values. As the pH value increased, the absorbance intensity distinctly decreased and the optimal pH were set as 4. The surface charges on the prepared nanocomposites would be altered with the pH-induced protonation of the carboxyl groups contained in the prepared nanocomposites.
To further verify the mechanism of DNA adsorption on MIL-88(NH2)-Fe@CNTs, zeta potentials of different components have been measured. As expected, the adsorption of DNA altered the surface charge of as-prepared nanocomposites, MIL-88(NH2)-Fe@CNTs is more negative (− 5.9 to − 23.8 eV) than MIL-88(NH2)-Fe (10.2 eV to − 18.7 eV), which is conductive to the binding between the catalysts and TMB substrate via electrostatic interaction (Fig. 8). Further, the comparison of fluorescence intensity of supernatant indicating MIL-88(NH2)-Fe@CNTs absorb more DNA, which shows MIL-88(NH2)-Fe@CNTs has a better affinity to DNA (Figure S4). As a result, the above results suggest that the introduction of CNTs not only improve the catalytic performance of MIL-88(NH2)-Fe via promoting the electron transfer, but also have an effect on the adsorption of DNA (Figure S4), which could be devoted to the flexible regulation of catalytic activities. Furthermore, in terms of zeta potentials Km and Vmax, were determined based on Michaelis-Menton equation as follows: V0 = Vmax [S]/(Km + [S]), where Km is regard as Michaelis constant, representing the affinity of enzyme or enzyme mimics to the substrates. Vmax refers to the maximum reaction velocity while [S] is the concentration of substrates. And we have fitted the Lineweaver–Burk curves with TMB as single variated factor, as well as H2O2, respectively (Figure S3). The calculated value of Km and Vmax are listed in Table 1, compared with MIL-88(NH2)-Fe, Vmax of MIL-88(NH2)-Fe@CNTs towards H2O2 is an order of magnitude higher than that of MIL-88(NH2)-Fe, while for TMB substrates, Vmax of MIL-88(NH2)-Fe@CNTs is comparable with MIL-88(NH2)-Fe, indicating the introduction of CNTs indeed enhance the catalytic reaction. In case of TMB substrates, Km value of MIL-88(NH2)-Fe@CNTs with ssDNA is 1.24 times lower than that of HRP, indicating that the our-prepared nanocomposites excellent affinity to TMB. Moreover, with H2O2 as substrates, the Km also decreased by 10.57 times compared to HRP. Importantly, the Vmax values for our-prepared nanocomposites even increased by four or five orders of magnitude. These results indicated that the MIL-88(NH2)-Fe@CNTs with ssDNA exhibit excellent affinity and catalytic velocity towards both TMB substrates and H2O2, which affirmed the above hypothesis.
4 Conclusions
In summary, we fabricated the MIL-88(NH2)-Fe@CNTs with controlled catalytic activity by ssDNA. Compared with pure MIL-88(NH2)-Fe nanorods, the introduction of CNTs enables the nanocomposites’ better affinity to DNA and enhancement of peroxidase-like activity, which achieves flexible regulation of their catalytic activity. It was because that the negatively charged ssDNA altered the surface charge properties of the nanocomposites to increase the electrostatic interaction between catalyst and substrates. Beyond that, the significant enhancement of the adsorption capacity towards ssDNA in the presence of CNTs was verified. We explore the switchable catalytic activity regulated by different length or sequence of ssDNA. Besides, the narrower bandgap of MIL-88(NH2)-Fe@CNTs is indeed conductive to accelerate electron transfer process, thus have a positive effect on the catalytic oxidation of substrates. Our study revealed that as-prepared nanocomposites with adsorbed ssDNA showed excellent affinity to substrates and high catalytic reaction rate according to Michaelis–Menten kinetics. More importantly, with the aid of good affinity of CNTs to DNA, our-prepared MIL-88(NH2)-Fe@CNTs exhibit enhanced DNA adsorption capacity, which pave the way of achieving ultrasensitive assays of various pollutants in actual environment.
References
Wei H, Wang E. Nanomaterials with enzyme-like characteristics (nanozymes): next-generation artificial enzymes. Chem Soc Rev. 2013;42(14):6060–93.
Xia XH, et al. Pd-Ir Core-Shell Nanocubes: a Type of Highly Efficient and Versatile Peroxidase Mimic. ACS Nano. 2015;9(10):9994–10004.
Gao LZ, et al. Intrinsic peroxidase-like activity of ferromagnetic nanoparticles. Nat Nanotechnol. 2007;2(9):577–83.
Kim CK, et al. Ceria Nanoparticles that can Protect against Ischemic Stroke. Angew Chem Int Ed. 2012;51(44):11039–43.
Manea F, et al. Nanozymes: gold-nanoparticle-based transphosphorylation catalysts. Angew Chem Int Ed. 2004;43(45):6165–9.
Song YJ, et al. Selective and quantitative cancer cell detection using target-directed functionalized graphene and its synergetic peroxidase-like activity. Chem Commun. 2011;47(15):4436–8.
Zhang EH, et al. Porous Ce3O4 hollow nanododecahedra for nonenzymatic glucose biosensor and biofuel cell. Biosens Bioelectron. 2016;81:46–53.
Huxford RC, Della Rocca J, Lin WB. Metal-organic frameworks as potential drug carriers. Curr Opin Chem Biol. 2010;14(2):262–8.
Li J, et al. Metal-organic framework-based materials: superior adsorbents for the capture of toxic and radioactive metal ions. Chem Soc Rev. 2018;47(7):2322–56.
Li WJ, et al. In Situ Growth of Metal-Organic Framework Thin Films with Gas Sensing and Molecule Storage Properties. Langmuir. 2013;29(27):8657–64.
Eddaoudi M, et al. Systematic design of pore size and functionality in isoreticular MOFs and their application in methane storage. Science. 2002;295(5554):469–72.
Chen BL, et al. A Luminescent Metal-Organic Framework with Lewis Basic Pyridyl Sites for the Sensing of Metal Ions. Angewe Chem Int Ed. 2009;48(3):500–3.
Nath I, Chakraborty J, Verpoort F. Metal organic frameworks mimicking natural enzymes: a structural and functional analogy. Chem Soc Rev. 2016;45(15):4127–70.
Gao W, et al. A competitive coordination-based CeO2 nanowire-DNA nanosensor: fast and selective detection of hydrogen peroxide in living cells and in vivo. Chem Commun. 2016;52(18):3643–6.
Qin FX, et al. Hemin@metal-organic framework with peroxidase-like activity and its application to glucose detection. Catal Sci Technol. 2013;3(10):2761–8.
Wang SQ, et al. Copper-based metal organic framework nanoparticles with peroxidase-like activity for sensitive colorimetric detection of Staphylococcus aureus. ACS Appl Mater Interfaces. 2017;9(29):24440–5.
Yang LZ, et al. An electrochemical sensor for H2O2 based on a new Co-metal-organic framework modified electrode. Sens Actuators B-Chem. 2015;215:489–96.
Jayasena SD. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin Chem. 1999;45(9):1628–50.
Chen M, et al. A novel aptamer- metal ions- nanoscale MOF based electrochemical biocodes for multiple antibiotics detection and signal amplification. Sens Actuators B-Chem. 2017;242:1201–9.
Fu Y, et al. DNA-Based platinum nanozymes for peroxidase mimetics. J Phys Chem C. 2014;118(31):18116–25.
Ning WY, et al. Imparting designer biorecognition functionality to metal-organic frameworks by a dna-mediated surface engineering strategy. Small. 2018;14(11):8.
Morris W, et al. Nucleic acid-metal organic framework (MOF) nanoparticle conjugates. J Am Chem Soc. 2014;136(20):7261–4.
Kempahanumakkagari S, et al. Biomolecule-embedded metal-organic frameworks as an innovative sensing platform. Biotechnol Adv. 2018;36(2):467–81.
Zhai YP, et al. Carbon materials for chemical capacitive energy storage. Adv Mater. 2011;23(42):4828–50.
Baughman RH, Zakhidov AA, de Heer WA. Carbon nanotubes–the route toward applications. Science. 2002;297(5582):787–92.
Xia Y, et al. A visible and colorimetric aptasensor based on DNA-capped single-walled carbon nanotubes for detection of exosomes. Biosens Bioelectron. 2017;92:8–15.
Peyvandi A, et al. Surface-modified graphite nanomaterials for improved reinforcement efficiency in cementitious paste. Carbon. 2013;63:175–86.
Shearer C, et al. Preparation and characterisation of vertically aligned single-walled carbon nanotube arrays on porous silicon. 2008; 7267.
Kim K, et al. Characterization of oxygen containing functional groups on carbon materials with oxygen K-edge X-ray absorption near edge structure spectroscopy. Carbon. 2011;49(5):1745–51.
Peng JB, et al. Catalytic effect of low concentration carboxylated multi-walled carbon nanotubes on the oxidation of disinfectants with Cl-substituted structure by a Fenton-like system. Chem Eng J. 2017;321:325–34.
Li X, et al. POMOF/SWNT Nanocomposites with Prominent Peroxidase-Mimicking Activity for L-Cysteine “On Off Switch” Colorimetric Biosensing. ACS Appl Mater Interfaces. 2019;11(18):16896–904.
Wang XT, et al. Integration of membrane filtration and photoelectrocatalysis on g-C3N4/CNTs/Al2O3 membrane with visible-light response for enhanced water treatment. J Membr Sci. 2017;541:153–61.
Horcajada P, et al. How linker’s modification controls swelling properties of highly flexible iron(III) dicarboxylates MIL-88. J Am Chem Soc. 2011;133(44):17839–47.
Xie DH, et al. Bifunctional NH2-MIL-88(Fe) metal-organic framework nanooctahedra for highly sensitive detection and efficient removal of arsenate in aqueous media. J Mater Chem A. 2017;5(45):23794–804.
Sun ZJ, Jiang JZ, Li YF. A sensitive and selective sensor for biothiols based on the turn-on fluorescence of the Fe-MIL-88 metal-organic frameworks-hydrogen peroxide system. Analyst. 2015;140(24):8201–8.
Bauer S, et al. High-throughput assisted rationalization of the formation of metal organic frameworks in the iron(III) aminoterephthalate solvothermal system. Inorg Chem. 2008;47(17):7568–76.
Pham MH, et al. Novel route to size-controlled Fe-MIL-88B-NH2 metal-organic framework nanocrystals. Langmuir. 2011;27(24):15261–7.
Zhang H, et al. Carbon nanotubes-incorporated MIL-88B-Fe as highly efficient Fenton-like catalyst for degradation of organic pollutants. Front Environ Sci Eng. 2019;13(2):11.
Tan B, et al. Fe3O4-AuNPs anchored 2D metal-organic framework nanosheets with DNA regulated switchable peroxidase-like activity. Nanoscale. 2017;9(47):18699–710.
Wang CH, Tang GG, Tan HL. Colorimetric determination of mercury(II) via the inhibition by ssDNA of the oxidase-like activity of a mixed valence state cerium-based metal-organic framework. Microchim Acta. 2018;185(10):8.
Liu M, et al. Stimuli-responsive peroxidase mimicking at a smart graphene interface. Chem Commun. 2012;48(56):7055–7.
Park KS, et al. Label-free colorimetric detection of nucleic acids based on target-induced shielding against the peroxidase-mimicking activity of magnetic nanoparticles. Small. 2011;7(11):1521–5.
Kim MI, Kim MS, Woo M-A, Ye Y, Kang KS, Lee J, Park HG. Highly efficient colorimetric detection of target cancer cells utilizing superior catalytic activity of graphene oxide–magnetic-platinum nanohybrids. Nanoscale. 2014;6(3):1529–36.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 21777012), the Program of Introducing Talents of Discipline to Universities (B13012), and the Program for Changjiang Scholars and Innovative Research Team in University (IRT_13R05).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
About this article
Cite this article
Sun, S., Fan, Y., Du, J. et al. CNT-Modified MIL-88(NH2)-Fe for Enhancing DNA-Regulated Peroxidase-Like Activity. J. Anal. Test. 3, 238–245 (2019). https://doi.org/10.1007/s41664-019-00111-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41664-019-00111-1