Abstract
Introduction
Accurate classification of somatic genetic alterations detected by next-generation sequencing (NGS) assays is of paramount importance to ensure the provision of high-quality clinical data. Clinical significance of variants can be assessed and tiered based on guidelines from the Association for Molecular Pathology (AMP), the American Society of Clinical Oncology, and the College of American Pathology for the interpretation of somatic sequence variants identified in cancer.
Methods
We sought to develop a formal structured approach for the classification of somatic variants in hematologic neoplasms, to account for both a variant’s clinical significance and its ability to drive tumorigenesis, by adapting elements from these existing guidelines. However, we additionally utilized key criteria from the American College of Medical Genetics/AMP standards for variant reporting to focus evaluation into specific categories of evidence and to gauge the effect of a given variant on tumorigenesis.
Results
The combined approach was applied to the annotation of 87 variants identified by a targeted NGS panel for myeloid neoplasms. In the application of our variant evaluation, we classified 2/87 variants as benign, 6/87 as likely benign, 56/87 as variants of unknown significance (VUS), 13/87 variants as likely pathogenic, and 10/87 variants as pathogenic.
Conclusion
Well-established oncogenic alterations were accurately classified as pathogenic. Although there is no defined benchmark for the remaining variants, drawing from two existing guidelines enabled the creation of a modified curation process for variant interpretation that emphasizes systematic review of relevant evidence.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
A systematic method for the classification of hematologic neoplasm (HN) variants identified by next-generation sequencing was newly developed. |
The framework combines elements from the Association for Molecular Pathology (AMP)/American Society of Clinical Oncology/College of American Pathology and American College of Medical Genetics/AMP guidelines to allow for the reporting of variants that (1) are clinically actionable, (2) drive tumorigenesis and may become clinically actionable, or (3) have implications for a hereditary predisposition to hematologic neoplasia or syndromes with hematological conditions. |
Consistency in classification of variants identified in HN is highly desirable and can be better achieved with an approach that accounts for some of the particular limitations of testing patients with HN. |
1 Introduction
Increased utilization of genetic testing has necessitated standardization in the interpretation of results to achieve interlaboratory consistency in reporting significant variants. Widespread integration of targeted panel, whole exome, whole genome, and transcriptomic sequencing by next-generation sequencing (NGS) methodologies has greatly expanded the amount of available clinical data requiring accurate and meaningful annotation and curation. In light of this challenge, the American College of Medical Genetics (ACMG) and the Association for Molecular Pathology (AMP) released updated guidelines for the interpretation of germline/constitutional sequence variants [1]. AMP, the American Society of Clinical Oncology (ASCO), and the College of American Pathology (CAP) similarly released guidelines for the interpretation of somatic sequence variants identified in cancer [2].
The ACMG/AMP guidelines for variant interpretation specify that they are not meant for use in the interpretation of somatic variants, whereas the AMP/ASCO/CAP guidelines provide a timely guidance for somatic variant curation, focusing on clinical impact from the therapeutic, diagnostic, and prognostic aspects. With this background, we found that the AMP/ASCO/CAP guideline classification schemes do not adequately address the evaluation of a given variant’s ability to drive tumorigenesis. In our clinical work-up of hematologic neoplasm (HN), we also found that the existing guidelines were more applicable to solid tumors than to HN. The main limiting factors include the current relatively limited availability of targeted therapies in HN; natural evolution from one specific HN entity to another (e.g., myelodysplastic syndrome to acute myeloid leukemia); and the not uncommon lack of a definitive diagnosis in cases under work-up, particularly for reference laboratories. Although consistency is desirable, both publications indicate that professional and clinical judgment may be incorporated into the variant curatioNAnnotation process based on individual circumstances. Furthermore, evaluation of the ACMG/AMP guidelines by the Clinical Genome Sequence Variant Interpretation (ClinGen SVI) Working Group indicated that the tiering of criteria was consistent with a Bayesian interpretation [3]. Finally, because detailed understanding of the pathogenesis of HN is continually evolving, consistent characterization of potential driver alterations is important to evaluate new information of clinical value. Based on these considerations, we devised a method for the interpretation of cancer sequence variants that incorporates elements from both guidelines.
2 Methods
2.1 Transcript Evaluation
It is generally necessary to check a variant’s effect in multiple transcripts, and a detailed approach was described for Mendelian disorders to narrow down transcripts of interest where possible [4]. To narrow down transcripts for somatic curation, we began by searching for clinically significant isoforms in Ensembl (www.ensembl.org or http://grch37.ensembl.org, depending on build) along with the closest reference sequence (RefSeq) match where available; then we recorded expression of all transcripts from the Genotype-Tissue Expression Portal (https://gtexportal.org) in tissues best related to the tumor site (e.g., whole blood and spleen). Although it is possible for neoplastic cells to alter transcript/isoform use, we excluded a given transcript in variant evaluation if it was not expected to produce a functional protein, had no evidence to suggest another role in hematopoietic processes, and/or had negligible expression in tissues of interest.
2.2 Criteria for the Interpretation of Sequence Variants
The criteria we use in tumor-derived variant evaluation are selected from those proposed by ACMG/AMP [1]. We removed certain nonapplicable criteria and repurposed those remaining—PVS1, PS1, PS3, PS4, PM1, PM2, PM4, PM5, PP3, BA1, BS1, BS3, BP3, BP4, and BP7—for use in the context of somatic curation in HN (Table 1). These are categorized into significant or supporting criteria in the following sections to demonstrate our approach.
2.2.1 Rules Deemed Not Applicable
Somatic panels include some genes (e.g., RUNX1, GATA2, CEBPA, ANKRD26, DDX41, ETV6, etc.) associated with Mendelian disorders. If the mechanism of germline disease matches the mechanism of tumorigenesis, utilizing some germline-specific criteria is possible (e.g., PP1/BS4—associated with familial co-segregation). However, we typically do not use PS2, PM3, PM6, PP1, PP2, PP4, PP5, BS2, BS4, BP1, BP2, BP5, and BP6.
2.2.2 Significant Criteria
2.2.2.1 Variant Type: BP7, PVS1, BP3, PM4
Certain types of variants imply a functional effect based on the predicted alteration to the messenger RNA (mRNA) and/or protein. The implied effect, when the mechanism of tumorigenesis is known and after considering relevant alternative transcripts/isoforms, contributes to the evidence supporting/refuting pathogenicity. Silent/synonymous variants typically represent a benign change, as do deep intronic and untranslated region (UTR) variants (BP7). However, it is important to rule out a splicing impact for silent variants by using in silico tools and consider the potential for a disrupted regulatory element (e.g., cis element, promoter, silencer/enhancer, etc.) or branch point for deep intronic/UTR variants.
Null variants, including nonsense variants, stop-gain frameshift variants, certain splicing variants, variants that alter the initiation codon, or out-of-frame single/multiexon deletions, are expected to result in loss of function of the protein product, which may be due to nonsense-mediated mRNA decay or a prematurely truncated protein (PVS1). The ClinGen SVI has enhanced the definition of the PVS1 criteria to address precautions and increase specificity, which we mimicked in our framework [5]. Stop-loss variants may result in an unstable product or a stable elongated product with neomorphic/enhanced properties or a dominant-negative effect. Thus, both null and stop-loss variants support a pathogenic effect.
In-frame deletions/duplications (indels) (BP3/PM4) may support either a benign or pathogenic effect depending on location and/or recurrent mutational pattern. However, it is difficult to determine whether missense variants favor pathogenicity or neutrality without additional information.
2.2.2.2 Minor Allele Frequency in the General (“Healthy”) Population: PM2, BA1, BS1
Assessing a variant’s minor allele frequency (MAF) in the general population approximates the frequency of the variant in “controls.” Public population databases that are commonly used to assess this include the Genome Aggregation Database (gnomAD, http://gnomad.broadinstitute.org/)—which includes new samples and high-quality samples from Exome Aggregation Consortium and 1000 Genomes Project—and the National Heart, Lung, and Blood Institute Exome Sequencing Project Exome Variant Server (ESP, http://evs.gs.washington.edu/EVS/) [6,7,8]. For most pathogenic variants, it is expected that they will be acquired and therefore absent from an unaffected general population (PM2). However, it is of note that these databases may not be able to exclude patients with clonal hematopoiesis of indeterminate potential (also called age-related clonal hematopoiesis) or subclinical diseases, including cancer. On the other hand, a MAF of ≥ 3% in the overall population or a continental population group is considered indicative of neutrality (BA1), whereas a MAF of 1–3% supports neutrality (BS1).
2.2.2.3 Frequency of Variant in the Cancer Population: PS4
The allele frequency of a variant in the cancer population is an ideal metric, but case-control studies are rare for any single variant in particular. Therefore, we counted confirmed somatic cases—using five as a conservative threshold when seen in the neoplastic group of interest—to serve as a proxy in support of pathogenicity. To determine the frequency of a variant in various tumor types, we used well-known databases: Catalogue of Somatic Mutations in Cancer (COSMIC, https://cancer.sanger.ac.uk/cosmic/) and cBioPortal for Cancer Genomics (cBioPortal, http://www.cbioportal.org/). We also aimed to be thorough by referring to additional databases when applicable and by checking for case reports across multiple transcripts, but we were careful to exclude overlapping cases across multiple databases and in the literature. Note that, if a gene was associated with a hereditary predisposition to cancer, we may have counted cases whether the variant’s origin has been confirmed as germline or is unknown.
2.2.2.4 Functional Study: PS3 and BS3
Provided a functional assay is reliable per the ACMG/AMP guidelines, functional data are considered to be one of the strongest pieces of evidence for supporting or refuting pathogenicity. Note that, if only one functional study was identified for a variant, we may still have counted this criterion but will have indicated the limitation in the report. Conflicting functional studies preclude use of these criteria.
2.2.2.5 Domain/Motif or Hotspot: PM1
If a variant occurs in a domain/motif with known or implicated protein function, it is more likely to support pathogenicity. However, instead of basing our decision on structural studies alone, we also incorporated the subregion residual variation intolerance score (http://www.subrvis.org/) and utilized gnomAD to approximate how many benign variants are reported in the domain [9]. In addition, if a variant occurs in a domain/motif that is highly enriched for pathogenic variants, it may suggest the domain/motif is a regional hotspot. Similarly, if a variant occurs at a residue where multiple cases have been reported (i.e., mutational hotspot), this also supports pathogenicity. The latter is typically assessed in our laboratory based on the number of reported cases in COSMIC/cBioPortal and the presence in Cancer Hotspot Databases (https://www.cancerhotspots.org/#/home and https://www.3dhotspots.org/#/home) [10,11,12].
2.2.2.6 Similar Pathogenic Variant: PS1 and PM5
The same protein change may be derived from various nucleotide changes; if one version of the variant has been shown to be likely pathogenic/pathogenic, it is highly likely that any DNA base alteration resulting in an identical protein change would create the same damaging effect (PS1) provided that the pathogenic mechanism of the established variant was not due to a splicing effect. On the other hand, if different missense variants occur at the same residue, the pathogenicity of one may not necessarily mean that the others have a deleterious effect. We gradated application of the criterion based on comparison of the physicochemical difference between the known pathogenic variant and the variant undergoing evaluation, as well as the overall differences between the residues in the wild-type and mutant proteins. Physicochemical difference can be both quantitatively and qualitatively measured by calculating Grantham distance and using software such as Alamut® Visual (SOPHiA GENETICS, Saint-Sulpice, Switzerland) [13]. Given a previously established missense variant is pathogenic, a new variant at the same residue that exhibits a large physicochemical difference compared with wild-type residue and a small physicochemical difference compared with the established mutant residue (e.g., both are hydrophobic) supports pathogenicity of the new variant (PM5). However, comparison of physicochemical difference may not be required if the variants are located at a known critical residue, such as a metal ion coordination site, cysteine disulfide bridge, etc. (PM5). See Sect. 2.2.3.2 for use of PM5 as supporting evidence.
2.2.2.7 Familial Segregation Analysis: PP1_strong/Moderate and BS4
If a variant in a tumor-derived sample occurs in a gene associated with predisposition to cancer, then the variant’s association with cancer or related phenotypes and its absence in unaffected relations supports pathogenicity (e.g., a RUNX1 variant only seen in relatives with either leukemia or thrombocytopenia) [14]. Richards et al. [1] emphasized the need for statistical evaluation to utilize this criterion but acknowledged that this may be difficult in a clinical laboratory setting. However, Jarvik and Browning [15] developed a method to quantify this criterion for application purposes. We utilized their method to determine how strongly familial data supported pathogenicity (PP1_strong/PP1_moderate). On the other hand, if the variant does not segregate with affected members in at least two unrelated families, this is sufficient to consider as a significant criterion for neutrality (BS4). See Sect. 2.2.3.3 for use of PP1 as supporting evidence.
2.2.3 Supporting Criteria
2.2.3.1 Computational Evidence: PP3 and BP4
Missense Tools
In silico tools approximate the functional effect of a variant largely based on species conservation and residue properties. Many predictors were developed by using training sets associated with Mendelian disease, but similar tools trained with cancer sets have been more recently evaluated [16, 17]. We employed consensus scoring across multiple tools to gauge support for pathogenicity or neutrality. Because research using existing missense predictors on somatic cancer variant datasets may reveal other useful tools, algorithms such as the Rare Exome Variant Ensemble Learner (REVEL) continue to be evaluated in our practice [18]. In our experience, evaluation of the REVEL meta-predictor has shown high concordance with currently employed single algorithmic applications.
Splicing Algorithms
In silico splice site/gene finders are utilized in curation given incomplete data regarding a variant’s effect on canonical or alternate mRNA splicing. Several papers provide data that may aid in defining how to interpret output from splicing algorithms, but we utilized a study from The National Genetics Reference Laboratory (Manchester, UK) for guidance [19,20,21]. Considering predictive scores and/or the magnitude of score changes at the canonical splice sites, the criterion was applied if consensus across multiple algorithms was present. However, impact on existing cryptic splice sites or their creation is more difficult to categorize with in silico tools given that experimental data on functional effects are typically absent, which makes validation and consequent application of the tools less reliable for these instances. Note that splicing algorithms alone may be supportive of neutrality if no or very minimal predictive changes are seen for intronic variants.
Protein Modeling
Understanding residue topography within 3D protein structure provides insight into residue–residue interactions, target binding in domains/motifs, effect on metal ion-coordinating sites, and how variants alter these conformations [22]. Pathogenicity may be inferred for a variant at a residue that is predicted to be involved in critical interactions or for a variant that alters the conformation of the protein by disrupting a domain/motif. However, neutrality is not automatically inferred for the inverse circumstance.
2.2.3.2 Similar Pathogenic Variant: PM5_supporting
When comparing different missense variants at the same residue, a previously established missense variant that is likely pathogenic is supportive of the new variant’s pathogenicity, unless the new substitution shows a small physicochemical difference between the wild-type and mutant residues. If the previously established missense variant is pathogenic, physicochemical differences that are less pronounced for a new alteration at the same residue may still support pathogenicity; as part of our proposed gradation based on physicochemical differences, we used this criterion as supporting information if there was (1) a large physicochemical difference between the wild-type and mutant residues or (2) a moderate physicochemical difference between the wild-type and mutant residues and a small physicochemical difference between the two mutant residues.
2.2.3.3 Familial Segregation Analysis: PP1
This supporting criterion is similar to the significant criterion, except that higher probabilities of segregation by chance would qualify as a supporting criterion for pathogenicity [15].
2.3 Variant Classification
Combining criteria should result in a classification in a five-tier system similar to the rules defined by Richards et al. [1]. We indicate the allowed combinations of criteria and how the corresponding evidence translates into a classification (Table 2). Note that evidence that does not count towards a criterion (Table 1) may still suggest whether a variant has some attributes favoring pathogenic/benign within the designated classification of variants of unknown significance (VUS).
2.4 Criteria to Determine the Clinical Significance of Sequence Variants
The primary purpose of tumor-derived variant evaluation is to determine whether identified variants can help diagnose/subtype a patient’s neoplasm, predict outcomes, and/or guide therapeutic choices. Therefore, even if a variant may not be an oncologic driver as indicated by the five-tier classification system, a context-specific and/or statistically significant association with outcomes may still exist (i.e., polymorphism associated with altered survival or therapy response). Li et al. [2] described how to weight evidence regarding clinical impact, with levels A and B evidence being stronger than levels C and D. We informally used this approach in curations but detailed the association in the report by explaining the aggregate evidence.
2.5 Sample Set
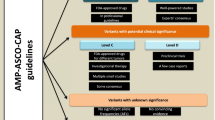
Our clinical laboratory utilizes a targeted 42-gene NGS panel for myeloid neoplasms. Variants are reviewed by a technical specialist and/or genetic counselor (GC) before a final evaluation and sign-out by a hematopathologist (Fig. 1). As this NGS panel has been in active high-volume use for over 6 years, most benign, likely benign, likely pathogenic, and pathogenic variants are known in new samples. To maintain laboratory process efficiency, these variants are analyzed and processed for hematopathologist sign-out by our technical specialists; the types of variants evaluated by GCs (i.e., novel and previously classified VUS) are focused on a smaller, more challenging subset. Because of this operational approach, we could not obtain variants in a prospective manner but instead compiled a set of 87 previously GC-curated variants seen in patient samples to demonstrate application of the aforementioned criteria and system.
Diagram of laboratory NGS workflow: Sequence calls (from variant call file, VCF) are initially reviewed by technical specialists. If there are no novel variants or previously classified VUS findings, the report is prepared by the technical specialist and passed on to a hematopathologist for review, edits, and final sign-out. However, when novel variants and/or previously classified VUS are identified in a sample, the case is first passed to a genetic counselor for in-depth review of the variant by the proposed classification scheme and then transferred to the hematopathologist for sign-out. This interpretive and curation/classification system optimizes efficiency while maintaining a high degree of consistency in our experience. CGSL Clinical Genome Sequencing Laboratory, NGS next-generation sequencing, VUS variants of unknown significance
3 Results and Discussion
In the application of our variant evaluation, we determined that 2/87 variants were benign, 6/87 were likely benign, 56/87 were VUS, 13/87 were likely pathogenic, and 10/87 were pathogenic (electronic supplementary material [ESM]-1). There is no absolute benchmark for many of these variants, and a majority of the variant classifications are notably VUS, but our modified approach allows us to anticipate whether a variant is favoring benign or pathogenic, enabling more efficient curation updates because the type of evidence that is needed to reach a likely benign/pathogenic classification is more apparent. The pathogenic variants were deemed correctly classified as they represent hotspot mutations that are well-established as oncogenic alterations in the peer-reviewed literature and cancer databases. For example, p.(Trp515Ser) in the MPL gene occurs at a hotspot residue (PM1), is not present in population databases (PM2), and has functional evidence supportive of ligand-independent activation in vitro (PS3). The variant has also been reported, yet not confirmed somatically, in ten or more myeloproliferative neoplasms (evidence favors pathogenicity, but it does not meet the rule for PS4). Finally, other W515 missense changes are described as pathogenic (PM5). As expected, the collective evidence indicated that the variant would be reportable (technically likely pathogenic but, based on our clinical judgment, upgraded to pathogenic), which also would align with AMP/ASCO/CAP guidelines (tier I based on its diagnostic and prognostic significance) [2].
However, our modified approach builds upon the existing guidelines by encouraging the reporting of variants that may have therapeutic/prognostic/diagnostic data available, although insufficient to meet any level of evidence defined in the AMP/ASCO/CAP guidelines [2]. This may be especially relevant for patients with refractory or relapsed disease. For example, p.(Gln61Leu) in KRAS occurs at a hotspot residue (PM1), is not present in population databases (PM2), and has functional evidence supportive of its transformative ability and extracellular signal-regulated kinase (ERK) activation (PS3). Furthermore, although c.182_183delinsTC observed in our data is a less common cause of this protein change than c.182A>T, both are reported as somatic variants in multiple neoplasms, with the latter including several hematological malignancies (PS1 based on c.182A>T, but the frequency of c.182_183delinsTC was insufficient to apply PS4). Per AMP/ASCO/CAP guidelines, this variant would likely be classified as tier III in the context of HN given KRAS alterations in HN are not associated with approved therapeutic strategies and prognostic significance is controversial for some leukemia subtypes [2]. However, a driver mutation such as this would be of relevance to report, especially because there are ongoing clinical trials for mitogen-activated protein kinase kinase (MEK)/phosphoinositide 3 kinase (PI3K)/protein kinase B (AKT)/mammalian target of rapamycin (mTOR) inhibitors with some supportive preclinical data or from a disease-monitoring perspective.
An additional use of the modified approach may be specific to testing performed without paired normal tissue, as the variant’s origin is not required to determine which set of guidelines should be applied for curation. For example, DDX41 is a gene that has recently been associated with germline predisposition to myeloid malignancies. The NM_016222.2(DDX41):c.415_418dup/p.(Asp140Glyfs*2) is a null variant (PVS1) that co-segregates with disease in multiple families (PP1_strong), which would yield a pathogenic classification (also likely per ACMG/AMP guidelines if treated as a germline variant) but would yield a tier III classification per AMP/ASCO/CAP guidelines if presumed to be a somatic variant. A tier III classification for this variant would diminish its impact, because—even though the variant itself is not associated with therapeutic significance—if it is confirmed to be of germline origin, family members who are carriers would be at increased clinical risk and inappropriate candidates for matched related donor bone marrow transplantation.
The aforementioned examples aim to show the potential to identify additional variants that may drive tumorigenesis and/or may eventually be of clinical significance. However, there are limitations to the evaluation of our model. First, we initially intended to prospectively classify variants identified in cases but, because the workflow funnels novel variants and VUS to the GCs who use this approach (Fig. 1), it may seem like the modified approach does not at first glance add value to the evaluation process. Consequently, we enriched the sample set with other variants identified in clinical samples to demonstrate the performance of the modified approach for additional unambiguous benign, likely benign, and pathogenic variants. While the sample set is no longer a prospective sampling, the expanded dataset has a representative distribution of variants in each class. A second limitation was related to our inability to compare the classifications from the modified approach to the definitive status (“ground truth”) of each variant, as the latter does not exist for every variant. As an alternative, variants were classified by the AMP/ASCO/CAP guidelines for comparison (ESM-1) [2].
In summary, all laboratories strive for consistent interpretation of genetic variants, internally and externally. To ensure such consistent interpretation, ACMG/AMP and AMP/ASCO/CAP released guidelines for the interpretation of germline/constitutional variants and somatic variants identified in cancer, respectively [1, 2]. By drawing from these two guidelines, our laboratory created a modified yet rigorous process for variant interpretation in HN NGS panels. Restriction of the criteria used in the ACMG/AMP guidelines with more specific requirements for many of the utilized criteria led to a conservative approach for classification of variants detected in our oncology panels without the requirement for matched normal tissue. After determining whether a variant has the potential to drive tumorigenesis or progression, the clinical significance of the variant can be assessed to incorporate into the report; this includes additional explanation for a likely pathogenic/pathogenic variant identified in a gene associated with a Mendelian disorder with recommendations to definitively confirm the variant in an appropriate germline sample (e.g., skin fibroblast sample), as it may affect general medical management and transplantation decisions when considering familial members. Although other laboratories may be using different frameworks, we hope that transparency will allow for critical review of our process such that improved consensus of somatic annotation is achieved with further modifications as needed.
References
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19:4–23.
Tavtigian SV, Greenblatt MS, Harrison SM, Nussbaum RL, Prabhu SA, Boucher KM, et al. Modeling the ACMG/AMP variant classification guidelines as a Bayesian classification framework. Genet Med. 2018;20:1054–60.
DiStefano MT, Hemphill SE, Cushman BJ, Bowser MJ, Hynes E, Grant AR, et al. Curating clinically relevant transcripts for the interpretation of sequence variants. J Mol Diagn. 2018;20:789–801.
Abou Tayoun AN, Pesaran T, DiStefano MT, Oza A, Rehm HL, Biesecker LG, et al. Recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion. Hum Mutat. 2018;39:1517–24.
Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–43.
Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91.
Consortium 1000 Genomes Project, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526:68–74.
Gussow AB, Petrovski S, Wang Q, Allen AS, Goldstein DB. The intolerance to functional genetic variation of protein domains predicts the localization of pathogenic mutations within genes. Genome Biol. 2016;17:9.
Chang MT, Asthana S, Gao SP, Lee BH, Chapman JS, Kandoth C, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol. 2016;34:155–63.
Chang MT, Bhattarai TS, Schram AM, Bielski CM, Donoghue MTA, Jonsson P, et al. Accelerating discovery of functional mutant alleles in cancer. Cancer Discov. 2018;8:174–83.
Gao J, Chang MT, Johnsen HC, Gao SP, Sylvester BE, Sumer SO, et al. 3D clusters of somatic mutations in cancer reveal numerous rare mutations as functional targets. Genome Med. 2017;9:4.
Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–4.
Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–405.
Jarvik GP, Browning BL. Consideration of cosegregation in the pathogenicity classification of genomic variants. Am J Hum Genet. 2016;98:1077–81.
Gnad F, Baucom A, Mukhyala K, Manning G, Zhang Z. Assessment of computational methods for predicting the effects of missense mutations in human cancers. BMC Genom. 2013;14(Suppl 3):S7.
Kerr ID, Cox HC, Moyes K, Evans B, Burdett BC, van Kan A, et al. Assessment of in silico protein sequence analysis in the clinical classification of variants in cancer risk genes. J Community Genet. 2017;8:87–95.
Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, et al. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet. 2016;99:877–85.
Houdayer C, Caux-Moncoutier V, Krieger S, Barrois M, Bonnet F, Bourdon V, et al. Guidelines for splicing analysis in molecular diagnosis derived from a set of 327 combined in silico/in vitro studies on BRCA1 and BRCA2 variants. Hum Mutat. 2012;33:1228–38.
Tang R, Prosser DO, Love DR. Evaluation of bioinformatic programmes for the analysis of variants within splice site consensus regions. Adv Bioinform. 2016;2016:5614058.
Hellen B. Splice site tools: a comparative analysis report. National Genetics Reference Laboratory | Manchester. 2009. http://www.ngrl.org.uk/Manchester/sites/default/files/publications/Informatics/NGRL_Splice_Site_Tools_Analysis_2009.pdf. Accessed 8 June 2021.
Kucukkal TG, Yang Y, Chapman SC, Cao W, Alexov E. Computational and experimental approaches to reveal the effects of single nucleotide polymorphisms with respect to disease diagnostics. Int J Mol Sci. 2014;15:9670–717.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Funding
No sources of funding were used to conduct this study or prepare this manuscript.
Conflict of interest/Competing interest
Nikita Mehta, Rong He, and David Viswanatha are or were employees of the Mayo Clinic (Department of Laboratory Medicine and Pathology, Molecular Hematology Laboratory) and have no conflicts of interest that are directly relevant to the content of this article. NM is currently employed at Memorial Sloan Kettering Cancer Center, New York, NY, USA.
Ethics approval
The Mayo Clinic Institutional Review Board (IRB) acknowledged that based on responses submitted to the Mayo Clinic IRB eSystem Human Subjects Research Wizard tool, and in accordance with the Code of Federal Regulations, 45 CFR 46.102, this activity did not require IRB review.
Consent to participate
Not applicable.
Consent for publication
The authors all read and approved the manuscript.
Availability of data and material
All variants evaluated in this study are provided in the ESM.
Code availability
Not applicable.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mehta, N., He, R. & Viswanatha, D.S. Internal Standardization of the Interpretation and Reporting of Sequence Variants in Hematologic Neoplasms. Mol Diagn Ther 25, 517–526 (2021). https://doi.org/10.1007/s40291-021-00540-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-021-00540-8