Abstract
Background and Objectives
Genetic studies have revealed that the regulated upon activation normal T-cell expressed and secreted (RANTES) −28C/G and −403G/A polymorphisms are associated with asthma risk, but contradictory findings have also been reported. Therefore, we undertook a meta-analysis on this topic.
Methods
The PubMed, Web of Science, China National Knowledge Infrastructure (CNKI), and Wanfang databases were used to identify relevant studies published in the medical literature from 1990 to March 26, 2014. Nine studies (containing 2,103 cases and 2,876 controls) investigated the −28C/G polymorphism, and 11 studies (including 2,015 cases and 1,909 controls) assessed the −403G/A polymorphism.
Results
The pooled results demonstrated that the −28C/G polymorphism was not associated with asthma risk in the overall populations (Caucasians, Asians, and a mixed population). However, in subgroup analysis according to age, the −28G allele was associated with an increased risk of asthma in children (odds ratio [OR] 1.27, 95 % confidence interval [CI] 1.03–1.57, P value for heterogeneity [P het] = 0.163, P value for the overall effect [P z] = 0.028). When we further stratified the studies performed in children on the basis of ethnicity, we found that the −28G allele was associated with an increased risk of asthma in Asian children (OR 1.28, 95 % CI 1.02–1.62, P het = 0.127, P z = 0.035), but not in Caucasian children (OR 1.20, 95 % CI 0.68–2.12, P het = 0.137, P z = 0.530). In subgroup analysis by asthma phenotype, no association between either atopic or non-atopic asthma and the −28C/G polymorphism was identified. For the −403G/A polymorphism, meta-analysis showed no association with asthma risk in the overall populations (Caucasians, Asians, and black people). In subgroup analyses by age, ethnicity, and asthma phenotype, we still did not find any association between the −403G/A polymorphism and asthma.
Conclusion
Current findings suggest an association between the −28G allele and asthma risk in Asian children but not in Caucasian children.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
The G allele of the regulated upon activation normal T-cell expressed and secreted (RANTES) −28C/G polymorphism was associated with asthma risk in Asian children but not in Caucasian children |
The RANTES −403G/A polymorphism was not associated with asthma risk |
1 Introduction
Inflammatory leukocyte recruitment to the lung is a prominent pathological feature of asthma [1]. This process is directed by small inflammatory soluble molecules known as chemokines. Chemokines represent a large family of 8–15 kDa chemotactic proteins, which are subdivided into four groups on the basis of the relative position of their first N-terminal cysteine residues and are named CXC, CC, CX3C, and C [2]. Experimental and clinical studies have demonstrated an important role for chemokines in orchestrating multiple aspects of the inflammatory response in asthma, including induction of leukocyte migration, inflammatory mediator release, and promotion of T helper-2 (Th2) inflammatory responses [3]. This accumulating evidence generates huge scientific interest in investigating the relationship between polymorphisms in chemokine genes and individual genetic predisposition to asthma.
The regulated upon activation normal T-cell expressed and secreted (RANTES) gene, which maps on 17 (q11.2–q12), is considered to be a candidate gene for asthma. Among identified single nucleotide polymorphisms (SNPs) in the RANTES gene, two mutations, a C to G substitution at position −28 (rs2280788) and a G to A substitution at position −403 (rs2107538) in the promoter region, are most frequently evaluated [4, 5]. The association between these two polymorphisms and asthma risk has been investigated in many genetic association studies; however, the results from individual studies remain inconsistent. Meta-analysis is a statistical technique that takes data from a number of independent studies and combines them to permit estimation of an overall effect. Up to now, several meta-analyses on these two SNPs and asthma risk have been performed [6–9], but their results still need to be assessed further. First, some eligible association studies were omitted. Second, studies presenting deviation from Hardy–Weinberg disequilibrium (HWE) in controls were not carefully evaluated, which may have resulted in bias and loss of precision in the estimation of the overall odds ratios (ORs). Third, some studies that had overlapped subjects were also included. Fourth, several new data sets have been published within the last three years. Thus, we conducted an updated meta-analysis to clarify the effect of the −28C/G and −403G/A polymorphisms in the RANTES gene on the risk of asthma.
2 Materials and Methods
2.1 Search Strategy
We used the PubMed, Web of Science, China National Knowledge Infrastructure (CNKI), and Wanfang databases to identify relevant studies published in the medical literature from 1990 to March 26, 2014. The search terms included “Regulated upon Activation Normal T-cell Expressed and Secreted”, “RANTES”, “CCL5”, “polymorphism”, “genetic variant”, “gene”, and “asthma”. The search results were limited to publications in English or Chinese. The reference lists in all relevant publications were also hand searched to identify additional studies missed by the database search.
2.2 Inclusion and Exclusion Criteria
Studies were included in our meta-analysis if they met the following criteria: (1) studies on human subjects; and (2) studies reporting genotype or allele frequencies of the −28C/G or −403G/A polymorphisms in subjects with asthma and non-asthma controls. Studies were excluded if one of the following criteria existed: (1) no control; (2) no usable data reported; or (3) studies presenting deviation from HWE in controls.
2.3 Data Extraction
Assessment for eligibility of studies and extraction of data was performed by two independent investigators. Disagreement over eligibility of a study was resolved through evaluation by a third reviewer and discussion until a consensus was reached. For each study, the following data were extracted: (1) first author; (2) publication year; (3) country in which the study was performed; (4) ethnicity; (5) phenotype of asthma; (6) age of subjects; (7) sample size; and (8) individual genotypes and genotype counts in order of preference.
2.4 Statistical Analyses
All statistical tests were performed with Stata version 11.0 software. We used raw data on genotype frequencies, without adjustment for calculation of the study-specific estimates of odds ratios (ORs) and 95 % confidence intervals (CIs). Between-study heterogeneity was assessed by the Q statistic (Cochrane’s Q), which is considered significant for P < 0.1. In the absence of heterogeneity, pooled estimation of ORs in each study was calculated by the fixed-effects model (Mantel–Haenszel methods) [10], while the random-effects model (DerSimonian and Laird’s method) was used to calculate pooled ORs in the case of no significant heterogeneity [11]. The significance of overall ORs was determined by the Z test, with P < 0.05 considered statistically significant. For assessing the relation of the −28C/G and −403G/A polymorphisms to asthma risk, the dominant, recessive, homozygote, and allelic comparison models were employed. Subgroup analyses were conducted according to ethnicity, age, and asthma phenotype, respectively. Publication bias was evaluated using funnel plots and Egger’s test. HWE in the controls for each study was tested by using a Web-based program (http://www.oege.org/software/hwe-mr-calc.html).
3 Results
3.1 Characteristics of the Included Studies
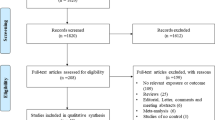
Figure 1 showed the process of identifying eligible studies. We retrieved 584 published studies, using our search criteria. After screening of the abstracts of these studies, 564 were excluded either because they were irrelevant, the record was a duplicate search result, or the study was not about human subjects. The remaining 20 studies were retrieved for more detailed evaluations, which excluded an additional six studies because no available data for genotypic distribution were reported [12, 13], the studies showed deviation from HWE in controls [14, 15], or the studies had overlapped subjects [16, 17], leaving 14 relevant publications to be included in the final analyses [18–31]. Table 1 shows the characteristics of the eligible studies included in this meta-analysis. Among them, nine studies (including a total of 2,103 cases and 2,876 controls) evaluated the −28C/G polymorphism [19, 21, 22, 24, 26–30], and 11 studies (containing 2,015 cases and 1,909 controls) assessed the −403G/A polymorphism [18–21, 23–27, 29, 31]. The study by Hizawa et al. [20] investigated both the −403G/A and −28C/G polymorphisms, but it had overlapped subjects with those from the study by Kaneko et al. [30], which assessed the −28C/G polymorphism with a larger sample size, so we excluded the data on the −28C/G polymorphism from the study by Hizawa et al. In addition, since the study by Liu et al. [30] presented a deviation from HWE in controls for the −28C/G polymorphism, we did not extract information on genotypic distribution for the −28C/G polymorphism from that study. In terms of ethnicity, seven studies were conducted in Asians [21–24, 26, 30, 31], four studies were performed in Caucasians [18, 19, 27, 29], one study was undertaken in black people [25], and one study was conducted in a mixed population [28]. Table 2 summarized the genotypic distribution in each study.
3.2 Association Between the −28C/G Polymorphism and Asthma Risk
All of the eligible studies for this SNP presented no deviation from HWE in controls. Pooling data provided no evidence of a relationship between this polymorphism and asthma risk in the overall populations (Caucasians, Asians, and a mixed group) in the dominant model (OR 1.22, 95 % CI 0.74–2.00, P value for heterogeneity [P het] = 0.276, P value for the overall effect [P z] = 0.117), recessive model (OR 1.21, 95 % CI 0.81–1.81, P het = 0.228, P z = 0.346), homozygote model (OR 1.25, 95 % CI 0.84–1.87, P het = 0.214, P z = 0.277), and allelic comparison model (OR 1.12, 95 % CI 0.98–1.28, P het = 0.222, P z = 0.094) [Fig. 2]. In subgroup analysis stratified by ethnicity, we did not find an association between the −28C/G polymorphism and asthma risk in Caucasians (506 cases and 583 controls) and Asians (1,497 cases and 1,811 controls), respectively (Table 2). In the stratified analysis based on age, we found that the G allele was associated with an increased risk of asthma in children (OR 1.27, 95 % CI 1.03–1.57, P het = 0.163, P z = 0.028) [Table 2]. However, no association was identified in the dominant, recessive, and homozygote models (Table 2). When we further stratified studies performed in children according to ethnicity, we found that the G allele was associated with an increased risk of asthma in Asian children (OR 1.28, 95 % CI 1.02–1.62, P het = 0.127, P z = 0.035) but not in Caucasian children (OR 1.20, 95 % CI 0.68–2.12, P het = 0.137, P z = 0.530). Because of the limited availability of published results, we were unable to evaluate this SNP in adults. Finally, we conducted subgroup analysis according to asthma phenotype. There was no significant association between either atopic or non-atopic asthma and the −28C/G polymorphism (Table 2).
3.3 No Association Between the −403G/A Polymorphism and Asthma Risk
No deviation from HWE was observed in any of the 11 independent studies. We found no association between the −403G/A polymorphism and asthma risk in the overall populations (Caucasians, Asians, and black people) in the dominant model (OR 1.04, 95 % CI 0.91–1.19, P het = 0.202, P z = 0.602), recessive model (OR 1.15, 95 % CI 0.93–1.42, P het = 0.823, P z = 0.206), homozygote model (OR 1.17, 95 % CI 0.93–1.46, P het = 0.836, P z = 0.183), and allelic comparison model (OR 1.05, 95 % CI 0.95–1.17, P het = 0.356, P z = 0.315) [Fig. 3]. In subgroup analysis stratified by ethnicity, no association between the −403G/A polymorphism and asthma risk was identified in Caucasians (626 cases and 625 controls) in the dominant model (OR 1.06, 95 % CI 0.82–1.36, P het = 0.037, P z = 0.676), recessive model (OR 1.31, 95 % CI 0.67–2.54, P het = 0.825, P z = 0.428), homozygote model (OR 1.33, 95 % CI 0.68–2.58, P het = 0.746, P z = 0.402), and allelic comparison model (OR 1.07, 95 % CI 0.86–1.34, P het = 0.077, P z = 0.528). We also did not find an association between the −403G/A polymorphism and asthma risk in Asians (1,351 cases and 1,153 controls), with similar ORs in these genetic models (Table 2). Age-specific analysis showed that the −403G/A polymorphism was not associated with asthma risk in either adults or children (Table 2). We further performed subgroup analysis according to asthma phenotype, finding that the −403G/A polymorphism was not associated with atopic or non-atopic asthma (Table 2).
3.4 Heterogeneity and Publication Bias
Table 3 shows between-study heterogeneity in detail. We found no heterogeneity in most analyses. We evaluated the −28C/G and −403G/A polymorphisms in the overall populations in the allelic comparison model for publication bias. Since publication bias was hard to detect when the number of studies were small, analysis for bias was not carried out in the subgroup analyses. The shapes of the funnel plots seemed to be symmetrical (Fig. 4). The P values for the −28C/G polymorphism (P = 0.437) and the −403G/A polymorphism (P = 0.511) were not significant.
4 Discussion
The main findings of this meta-analysis were as follows: (1) there was no significant association between the RANTES −28C/G SNP and asthma risk in the overall populations; (2) in subgroup analysis according to age and ethnicity, the G allele of the −28C/G polymorphism was associated with an increased risk of asthma in Asian children but not in Caucasian children; and (3) no association between the RANTES −403G/A polymorphism and asthma risk was identified in the overall populations. In subgroup analysis based on ethnicity, age, and asthma phenotype, we still did not find an association.
As increased infiltration of inflammatory cells into the lung is closely associated with development of asthma, the involvement of RANTES in asthma has attracted considerable interest. In vitro and animal studies have demonstrated that RANTES is a major chemotactic factor produced during the asthmatic response and contributes to exacerbation of allergic airway inflammation [32–34]. Increased levels of RANTES were observed in bronchial lavage fluid from patients with active asthma [35]. In addition, significantly higher plasma levels of RANTES were also detected in asthmatic patients during acute attacks [36]. Thus, the RANTES gene is thought to be a candidate gene for asthma susceptibility. The −28C/G and −403G/A polymorphisms are two functional variants that increase transcriptional activity and subsequent RANTES expression in human cell lines. In this meta-analysis, we first identified a positive association between the G allele of the −28C/G variant and asthma risk in Asian children but not in Caucasian children. It was reported that the −28G allele was associated with elevated RANTES messenger RNA and protein expression, an increased blood eosinophil count, and a higher degree of bronchial hyperresponsiveness. Therefore, it may increase the individual genetic predisposition to develop asthma. The discrepancy of the −28G allele association between Asian children and Caucasian children indicated that this SNP may have different effects on asthma risk according to the ethnic genetic background. In subgroup analysis according to age, because of the limited availability of published results, we were unable to investigate the association between the −28G allele and asthma in adults. We expect that as more studies become available, an accurate estimation of the relationship between the −28G allele and asthma in adults will be obtained.
Two previous meta-analysis obtained similar findings for the −28C/G polymorphism [7, 8]. With a smaller number of subjects, a meta-analysis by Fang et al. [7] found an association between the −28G allele and asthma risk in Asians. An ethnicity-specific meta-analysis [8] reported an association between the −28C/G polymorphism and asthma in Chinese populations. However, neither of those two meta-analyses further stratified Asian subjects according to age to assess the relationship between the −28C/G polymorphism and asthma. Therefore, they were unable to simultaneously evaluate the impact of ethnicity and age, which were considered to be the two most important factors accounting for heterogeneity in the RANTES association with asthma risk. The Lu et al. meta-analysis [9] focused on the association between the RANTES polymorphisms and the risk of pediatric asthma. However, because of the small number of subjects, that meta-analysis did not identify a significant association between the RANTES polymorphisms and pediatric asthma in Asians and Caucasians. The Zhang et al. meta-analysis [6] summarizing studies on the relation of the two RANTES SNPs, published up until November 2009, found an association between the −403G/A polymorphism and atopic asthma. However, four new studies have been published since 2009, dramatically increasing the numbers of asthma patients and controls with relevant genetic information [28–31]. In our updated meta-analysis including the newly available published studies, we did not find an association between either atopic or non-atopic asthma and the two RANTES polymorphisms.
This meta-analysis represents an updated and comprehensive review of the literature on the two most studied RANTES polymorphisms and asthma risk. There were some advantages in this study. Besides performing pooled analysis in the overall populations, we also conducted subgroup analyses based on ethnicity, age, and asthma phenotype. In order not to miss any potential literatures, besides electronic databases, we manually searched the reference lists in all relevant publications. We excluded studies containing overlapped subjects with a smaller sample size. For example, the Tölgyesi study [17] was excluded from our analysis in that it had overlapped subjects with those of the Szalai study [19]. The study by Huang [16] was also excluded for a similar reason. In addition, we excluded studies deviating from HWE in controls. Two studies were excluded for this reason [14, 15]. In terms of between-study heterogeneity and publication bias, most analyses showed no or very low heterogeneity, and there was no evidence of publication bias. Therefore, the results of our study were reliable to some extent.
Some limitations of this meta-analysis should be mentioned. First, the small sample size in some of the included studies might have influenced the statistical power to better evaluate the association between the −28C/G and −403G/A polymorphisms and asthma risk, especially in subgroup analysis. Second, because of the limited availability of published results, gene–gene and gene–environment interactions were not analyzed in this meta-analysis. Third, we evaluated only the −28C/G and 403G/A polymorphisms. It is also possible that other polymorphisms in the RANTES gene influence the risk of asthma.
5 Conclusion
The results of our meta-analysis demonstrate that the G allele of the RANTES −28C/G polymorphism is associated with an increased risk of asthma in Asian children but not in Caucasian children. There is no association between the RANTES −403G/A polymorphism and asthma risk.
References
Fireman P. Understanding asthma pathophysiology. Allergy Asthma Proc. 2003;24(2):79–83.
Zlotnik A, Yoshie O, Nomiyama H. The chemokine and chemokine receptor superfamilies and their molecular evolution. Genome Biol. 2006;7(12):243.
Garcia G, Godot V, Humbert M. New chemokine targets for asthma therapy. Curr Allergy Asthma Rep. 2005;5(2):155–60.
al Sharif F, Ollier WE, Hajeer AH. A rare polymorphism at position −28 in the human RANTES promoter. Eur J Immunogenet. 1999;26(5):373–4.
Hajeer AH, al Sharif F, Ollier WE. A polymorphism at position −403 in the human RANTES promoter. Eur J Immunogenet. 1999;26(5):375–6.
Zhang YG, Huang J, Zhang J, Li XB, He C, Xiao YL, Tian C, Wan H, Zhao YL, Tsewang YG, Fan H. RANTES gene polymorphisms and asthma risk: a meta-analysis. Arch Med Res. 2010;41(1):50–8.
Fang Q, Wang F, Zhao D. Association between regulated upon activation, normal T cells expressed and secreted (RANTES) −28C/G polymorphism and asthma risk—a meta-analysis. Int J Med Sci. 2010;7(1):55–61.
Li X, Zhang Y, Zhang J, Xiao Y, Huang J, Tian C, He C, Deng Y, Yang Y, Fan H. Asthma susceptible genes in Chinese population: a meta-analysis. Respir Res. 2010;11:129.
Lu YM, Cao LF, Li YQ, Li C. RANTES gene polymorphisms and risk of pediatric asthma: a meta-analysis. Exp Ther Med. 2012;4(5):918–22.
Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–48.
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–88.
Liu CH, Chen H, Hu LP, Fu J, Zhang HQ, Zhang JL, Chen ZL, Chen YZ. Association between the genetic polymorphism of chemokine genes and asthma in Chinese children. Zhonghua Er Ke Za Zhi. 2005;43(6):462–3 (in Chinese).
Huo J, Liu QH, Hua L, Bao YX. Association of single nucleotide polymorphisms in RANTES and eotaxin-3 genes with childhood asthma. Shanghai Jiao Tong Da Xue Xue Bao (Yi Xue Ban). 2010;30(2):129–31 (in Chinese).
Al-Abdulhadi SA, Helms PJ, Main M, Smith O, Christie G. Preferential transmission and association of the −403 G–>A promoter RANTES polymorphism with atopic asthma. Genes Immun. 2005;6(1):24–30.
Lachheb J, Chelbi H, Hamzaoui K, Hamzaoui A. Association between RANTES polymorphisms and asthma severity among Tunisian children. Hum Immunol. 2007;68(8):675–80.
Huang JL. Asthma severity and genetics in Taiwan. J Microbiol Immunol Infect. 2005;38(3):158–63.
Tölgyesi G, Keszei M, Ungvári I, Nagy A, Falus A, Szalai C. Involvement of TNFalpha −308A promoter polymorphism in the development of asthma in children infected with Chlamydophila pneumoniae. Pediatr Res. 2006;60(5):543–8.
Fryer AA, Spiteri MA, Bianco A, Hepple M, Jones PW, Strange RC, Makki R, Tavernier G, Smilie FI, Custovic A, Woodcock AA, Ollier WE, Hajeer AH. The −403 G–>A promoter polymorphism in the RANTES gene is associated with atopy and asthma. Genes Immun. 2000;1(8):509–14.
Szalai C, Kozma GT, Nagy A, Bojszkó A, Krikovszky D, Szabó T, Falus A. Polymorphism in the gene regulatory region of MCP-1 is associated with asthma susceptibility and severity. J Allergy Clin Immunol. 2001;108(3):375–81.
Hizawa N, Yamaguchi E, Konno S, Tanino Y, Jinushi E, Nishimura M. A functional polymorphism in the RANTES gene promoter is associated with the development of late-onset asthma. Am J Respir Crit Care Med. 2002;166(5):686–90.
Yao TC, Kuo ML, See LC, Chen LC, Yan DC, Ou LS, Shaw CK, Huang JL. The RANTES promoter polymorphism: a genetic risk factor for near-fatal asthma in Chinese children. J Allergy Clin Immunol. 2003;111(6):1285–92.
Wang LJ, Li YR, Chen JH, Cui TP, Wu JM. Polymorphism of regulated upon activation, normal T cell expressed and secreted promoter region −28 position in Chinese allergic asthmatic children. Zhonghua Jie He He Hu Xi Za Zhi. 2004;27(6):394–7 (in Chinese).
Leung TF, Tang NL, Lam CW, Li AM, Fung SL, Chan IH, Wong GW. RANTES G-401A polymorphism is associated with allergen sensitization and FEV1 in Chinese children. Respir Med. 2005;99(2):216–9.
Liu M, Li HL, Huang YK, Chen YH, Liu H, Jin P. The SNPs of chemokine RANTES promoter in children with asthma. Zhongguo You Sheng Yu Yi Chuan Za Zhi. 2005;13(11):20–3.
Moissidis I, Chinoy B, Yanamandra K, Napper D, Thurmon T, Bocchini J Jr, Bahna SL. Association of IL-13, RANTES, and leukotriene C4 synthase gene promoter polymorphisms with asthma and/or atopy in African Americans. Genet Med. 2005;7(6):406–10.
Sohn MH, Kim SH, Kim KW, Jee HM, Park HS, Kim KE. RANTES gene promoter polymorphisms are associated with bronchial hyperresponsiveness in Korean children with asthma. Lung. 2008;186(1):37–43.
Muro M, Marín L, Torio A, Pagan JA, Alvarez-López MR. CCL5/RANTES chemokine gene promoter polymorphisms are not associated with atopic and nonatopic asthma in a Spanish population. Int J Immunogenet. 2008;35(1):19–23.
Murk W, Walsh K, Hsu LI, Zhao L, Bracken MB, Dewan AT. Attempted replication of 50 reported asthma risk genes identifies a SNP in RAD50 as associated with childhood atopic asthma. Hum Hered. 2011;71(2):97–105.
Nahas R, Fakhoury HM, Chmaisse HN, Makki RF. Study of the association between −403G/A and −28C/G RANTES gene polymorphisms and asthma in Lebanon. Ann Thorac Med. 2012;7(1):16–20.
Kaneko Y, Masuko H, Sakamoto T, Iijima H, Naito T, Yatagai Y, Yamada H, Konno S, Nishimura M, Noguchi E, Hizawa N. Asthma phenotypes in Japanese adults—their associations with the CCL5 and ADRB2 genotypes. Allergol Int. 2013;62(1):113–21.
Liu Q, Hua L, Fang D, Lin Q, Zhu Y, Gan X, Bao Y. Interleukin-13 and RANTES polymorphisms in relation to asthma in children of Chinese Han nationality. Asian Pac J Allergy Immunol. 2013;31(3):247–52.
Lukacs NW1, Standiford TJ, Chensue SW, Kunkel RG, Strieter RM, Kunkel SL. C-C chemokine-induced eosinophil chemotaxis during allergic airway inflammation. J Leukoc Biol. 1996;60(5):573–8.
Pan ZZ, Parkyn L, Ray A, Ray P. Inducible lung-specific expression of RANTES: preferential recruitment of neutrophils. Am J Physiol Lung Cell Mol Physiol. 2000;279(4):L658–66.
John AE, Berlin AA, Lukacs NW. Respiratory syncytial virus-induced CCL5/RANTES contributes to exacerbation of allergic airway inflammation. Eur J Immunol. 2003;33(6):1677–85.
Alam R, York J, Boyars M, Stafford S, Grant JA, Lee J, Forsythe P, Sim T, Ida N. Increased MCP-1, RANTES, and MIP-1alpha in bronchoalveolar lavage fluid of allergic asthmatic patients. Am J Respir Crit Care Med. 1996;153(4 Pt 1):1398–404.
Chihara J1, Yasuba H, Tsuda A, Urayama O, Saito N, Honda K, Kayaba H, Yamashita T, Kurimoto F, Yamada H. Elevation of the plasma level of RANTES during asthma attacks. J Allergy Clin Immunol. 1997;100(6 Pt 2):S52–5.
Acknowledgments
This work was supported in part by a research grant (No. 2013GXNFAA019206) from the Natural Science Foundation of Guangxi Zhuang Autonomous Zone, P. R. China. The funding sources had no role in the study design; the collection, analysis, and interpretation of data; the writing of the report; or the decision to submit the article for publication.
Conflicts of interest
The authors declare no competing interests.
Author information
Authors and Affiliations
Corresponding author
Additional information
Z.-K. Xie and H. Zhao contributed equally to this research.
Rights and permissions
About this article
Cite this article
Xie, ZK., Zhao, H., Huang, J. et al. The Regulated upon Activation Normal T-cell Expressed and Secreted (RANTES) −28C/G and −403G/A Polymorphisms and Asthma Risk: A Meta-analysis. Mol Diagn Ther 18, 523–531 (2014). https://doi.org/10.1007/s40291-014-0112-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-014-0112-5