Abstract
Background
Voriconazole, a first-line antifungal drug, exhibits nonlinear pharmacokinetics (PK), together with large interindividual variability but a narrow therapeutic range, and markedly inhibits cytochrome P450 (CYP) 3A4 in vivo. This causes difficulties in selecting appropriate dosing regimens of voriconazole and coadministered CYP3A4 substrates.
Objective
This study aimed to investigate the metabolism of voriconazole in detail to better understand dose- and time-dependent alterations in the PK of the drug, to provide the model basis for safe and effective use according to CYP2C19 genotype, and to assess the potential of voriconazole to cause drug–drug interactions (DDIs) with CYP3A4 substrates in more detail.
Methods
In vitro assays were carried out to explore time-dependent inhibition (TDI) of CYP3A4 by voriconazole. These results were combined with 93 published concentration–time datasets of voriconazole from clinical trials in healthy volunteers to develop a whole-body physiologically based PK (PBPK) model in PK-Sim®. The model was evaluated quantitatively with the predicted/observed ratio of the area under the plasma concentration–time curve (AUC), maximum concentration (Cmax), and trough concentrations for multiple dosings (Ctrough), the geometric mean fold error, as well as visually with the comparison of predicted with observed concentration–time datasets over the full range of recommended intravenous and oral dosing regimens.
Results
The result of the half maximal inhibitory concentration (IC50) shift assay indicated that voriconazole causes TDI of CYP3A4. The PBPK model evaluation demonstrated a good performance of the model, with 71% of predicted/observed aggregate AUC ratios and all aggregate Cmax ratios from 28 evaluation datasets being within a 0.5- to 2-fold range. For those studies reporting CYP2C19 genotype, 89% of aggregate AUC ratios and all aggregate Cmax ratios were inside a 0.5- to 2-fold range of 44 test datasets. The results of model-based simulations showed that the standard oral maintenance dose of voriconazole 200 mg twice daily would be sufficient for CYP2C19 intermediate metabolizers (IMs; *1/*2, *1/*3, *2/*17, and *2/*2/*17) to reach the tentative therapeutic range of > 1–2 mg/L to < 5–6 mg/L for Ctrough, while 400 mg twice daily might be more suitable for rapid metabolizers (RMs; *1/*17, *17/*17) and normal metabolizers (NMs; *1/*1). When the model was integrated with independently developed CYP3A4 substrate models (midazolam and alfentanil), the observed AUC change of substrates by voriconazole was inside the 90% confidence interval of the predicted AUC change, indicating that CYP3A4 inhibition was appropriately incorporated into the voriconazole model.
Conclusions
Both the in vitro assay and model-based simulations support TDI of CYP3A4 by voriconazole as a pivotal characteristic of this drug’s PK. The PBPK model developed here could support individual dose adjustment of voriconazole according to genetic polymorphisms of CYP2C19, and DDI risk management. The applicability of modeling results for patients remains to be confirmed in future studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
A whole-body physiologically based pharmacokinetic (PBPK) model of voriconazole incorporating time-dependent inhibition (TDI), specifically mechanism-based inhibition (MBI) of cytochrome P450 (CYP) 3A4, was successfully developed to accurately capture the time- and dose-dependent alterations of voriconazole PK for different CYP2C19 genotypes. |
Model-based simulations could (1) elaborate potential exposure-equivalent dosing regimens for CYP2C19 genotype groups; (2) assess the dynamic inhibition of CYP3A4 by voriconazole in the liver and small intestine; and (3) predict DDIs between voriconazole and other CYP3A4 substrates. |
1 Introduction
Voriconazole is an essential drug in the treatment of severe fungal infections due to its activity against a wide range of clinically relevant fungal pathogens, including the most commonly occurring species of the genera Aspergillus and Candida, and some emerging fungi, such as Scedosporium and Fusarium species [1]. Moreover, voriconazole is well-established as first-line therapy for patients with invasive aspergillosis [2,3,4]. However, the drug exhibits nonlinear PK with large interindividual and intraindividual variability [5, 6], which causes difficulties for clinicians when choosing appropriate dosing regimens to target its narrow therapeutic range, especially in the case of high doses in severe infections, or for long-term treatments [7].
While underexposure of voriconazole may decrease efficacy, overexposure increases the risk primarily for neural and hepatic toxicity [8, 9]. Until now, no universally applicable therapeutic range has been established. In 2013, two Japanese societies recommended voriconazole trough concentrations for multiple dosings (Ctrough) of 1–2 mg/L to 4–5 mg/L [10], while in 2014 the British Society for Medical Mycology recommended Ctrough of 1 mg/L to 4–6 mg/L [11]. In 2017, according to the Third Fungal Diagnosis and Management of Aspergillus diseases Clinical Guideline, a Ctrough range of 1–5.5 mg/L was considered adequate for most patients with voriconazole prophylaxis or treatment, while the recommended range for patients with severe infections was 2–6 mg/L [4]. In 2018, the Chinese Pharmacological Society recommended a range of 0.5–5 mg/L [12]. Thus, in the present project, we selected lower and upper Ctrough of > 1–2 mg/L and < 5–6 mg/L, respectively.
Voriconazole is extensively metabolized via the cytochrome P450 (CYP) enzymes CYP2C19 and CYP3A4 [13], slightly by CYP2C9 and flavin-containing monooxygenase (FMO) [14], while < 2% is excreted renally as the parent drug [15,16,17]. The main metabolite in plasma was reported as voriconazole N-oxide, accounting for 72% of circulating metabolites [1]. However, Geist et al. found that voriconazole N-oxide and its conjugates excreted in urine within 12 h postdose during steady-state only accounted for 1% of the dose, while excretion of other metabolites, i.e. dihydroxy fluoropyrimidine–voriconazole and hydroxy fluoropyrimidine–voriconazole, together with their conjugates, accounted for 14% and 3% of the dose, respectively [17]. This was in agreement with another study where the major metabolite excreted in urine over 96 h was dihydroxy fluoropyrimidine–voriconazole, accounting for 13% of the dose of voriconazole [18]. Therefore, it seems reasonable to also consider dihydroxy-fluoropyrimidine voriconazole and hydroxy-fluoropyrimidine voriconazole as major metabolites of voriconazole, although both have low plasma concentrations due to their high renal clearances, which was reported to be approximately 150- and 55-fold higher, respectively, than that of voriconazole N-oxide [17]. However, two other groups found that the main metabolite of voriconazole excreted in urine within 48 h after administration was voriconazole N-oxide, accounting for 10–21% of the dose [15, 16]. The discrepancies between the studies may be explained by the respective length of urine collection periods, together with the different elimination half-life of the metabolites and a potential time-dependent inhibition (TDI) of CYP3A4. Thus, both fluoropyrimidine hydroxylation and N-oxidation pathways were considered as the main metabolic pathways, mainly mediated by CYP3A4 and CYP2C19, as shown in Fig. 1.
Metabolic pathway for voriconazole. *Indirect evidence from different CYP2C19 genotype groups [18]. CYP cytochrome P450, FMO flavin-containing monooxygenase
Genetic polymorphisms of CYP2C19 are a major source for interindividual variability, as reflected by threefold higher maximum concentration (Cmax) values and two- to fivefold higher area under the plasma concentration–time curve (AUC) values in CYP2C19 poor metabolizers (PMs) compared with those in normal metabolizers (NMs) or rapid metabolizers (RMs) [7, 19, 20].
Furthermore, voriconazole is also an inhibitor of CYP3A4 and CYP2C19 [21]. In vitro, voriconazole inhibition constant (Ki) for the competitive inhibition of CYP3A4-mediated metabolism of midazolam was reported to range from 0.15 to 0.66 µM [21, 22], indicating potent inhibition. In agreement with the in vitro results, the AUC of midazolam was considerably increased to 940% and 353% by oral and intravenous coadministration of therapeutic doses of voriconazole in vivo, respectively [23]. Furthermore, voriconazole was reported to mediate ‘autoinhibition’ of CYP3A4 activity in vivo [15, 24]. In addition, to properly describe the respective processes concerning enzyme inhibition by voriconazole in vivo, ‘TDI’ and ‘autoinhibition’, respectively, of voriconazole were integrated into the nonlinear mixed-effects models reported by Friberg et al. and Kim et al., respectively [25, 26].
Therefore, we investigated the inhibition of voriconazole and its metabolite voriconazole N-oxide on CYP3A4 and CYP2C19 in vitro. Based on the in vitro assay results, a whole-body physiologically based pharmacokinetic (PBPK) model of voriconazole incorporating CYP3A4 TDI was then developed to describe dose- and time-dependent PK in the different CYP2C19 genotypes. Finally, model-based simulations were carried out to (1) elaborate potentially exposure-equivalent dosing regimens for CYP2C19 genotype groups; (2) assess the dynamic inhibition of CYP3A4 by voriconazole in the liver and small intestine; and (3) further evaluate drug–drug interactions (DDIs) between voriconazole and other CYP3A4 probe substrates. An early stage of this work has been presented at the Population Approach Group in Europe conference [27].
2 Methods
2.1 In Vitro Assay for Inhibition of Cytochrome P450 (CYP) CYP2C19 and CYP3A4
The in vitro assay for inhibition of human CYP2C19 and CYP3A4 by voriconazole and its metabolite voriconazole N-oxide, together with the respective measurements and data analysis, were carried out according to the methods described in the electronic supplementary materials (ESM).
2.2 Model Development
The PBPK model for voriconazole was developed by combining bottom-up and top-down approaches. An extensive literature search was performed to obtain (a) drug physiochemical properties; (b) PK parameters describing absorption, distribution, metabolism and excretion processes; and (c) clinical studies of intravenous and oral administration of voriconazole to healthy subjects with different dosing regimens. The clinical studies were screened and selected according to the following criteria: (1) intravenous or oral administration of voriconazole; (2) healthy volunteers; (3) plasma concentration–time datasets of voriconazole were available; and (4) articles published in English. The training dataset for model development was selected based on (1) the information required for each step of model development; (2) the parameters need to be optimized; (3) the number of studies available; and (4) the informative content of datasets for individual studies (genotype groups, dosing regimens, and routes of administration), as shown in Fig. 2. Except datasets required and used for model development, all the remaining clinical trial datasets were utilized for model evaluation. The contribution of training datasets containing aggregate data from each clinical study was weighted equally to enable incorporation of some clinical studies that provided important information but did not report standard deviation (SD) or another measure of variability. Individual concentration–time datasets were pooled according to genotype groups, with the contribution of each individual dataset being weighted equally.
Workflow of voriconazole PBPK model development and evaluation. The PK datasets used to select the distribution model were also utilized to optimize Vmax and kinact for CYP3A4. There were 21 PK datasets for model development and 72 for model evaluation in total. ADME absorption, distribution, metabolism, elimination, PK pharmacokinetics, TDI time-dependent inhibition, PMs poor metabolizers, DDIs drug–drug interactions, PBPK physiologically based pharmacokinetic, CYP cytochrome P450, Vmax maximum velocity, kinact maximum inactivation rate constant
The modeling software PK-Sim® (version 7.3.0, part of the Open Systems Pharmacology (OSP) suite) was used for model development, which consists of a system- and drug-dependent component. System-dependent physiological parameters (organ volumes, blood flow rates, hematocrit, etc.) were provided in PK‐Sim® with the small molecule model [28,29,30]. Demographic characteristics of subjects were taken from each clinical study. Drug-specific physicochemical properties were obtained from the literature. Organ–plasma partition coefficients were determined using the Poulin and Theil method based on both the literature [31] and the best overlap between observed and predicted concentration–time datasets.
The workflow of model development is presented in Fig. 2. For model development, the simplifying assumption was made that the metabolism of voriconazole is mediated exclusively by CYP3A4 and CYP2C19; the minor contributions of CYP2C9, FMOs, and unchanged renal elimination of voriconazole were neglected [13, 16]. Tissue expression distribution of enzymes was provided by the PK-Sim® expression database based on reverse transcription-polymerase chain reaction (RT-PCR) profiles [32], together with the reference value of 4.32 µmol CYP3A4 and 0.76 µmol CYP2C19 per liter of liver tissue [33]. The relative CYP2C19 expression for different genotypes was obtained based on the CYP2C19 protein content ratio in genotype-defined pooled human liver microsomes [34]. The metabolism process of voriconazole was described using Michaelis–Menten kinetics [35]. As reported by Damle et al. [31], Km for CYP3A4 and CYP2C19 was set to 15 and 3.5 µM, respectively, and maximum velocity (Vmax) for CYP2C19 was fixed to 1.19 pmol/min/pmol. Vmax for CYP3A4 was optimized based on the concentration–time datasets in CYP2C19 PMs [18], with the assumption that only CYP3A4 contributes to the metabolism of voriconazole in PMs. TDI was integrated into the model assuming that it reflects mechanism-based inhibition (MBI), with Eq. S4 in the ESM based on the in vitro inactivity assay results of KI (the inhibition concentration when reaching half of kinact). The other parameter kinact (maximum inactivation rate constant) was optimized based on concentration–time curves after multiple intravenous administrations [36], since the in vitro derived kinact parameter value led to an overprediction of midazolam AUCs when evaluating the voriconazole–midazolam DDI studies.
The specific intestinal permeability was optimized based on the studies, including both intravenous and oral administration of voriconazole [6, 37, 38]. The dissolution of the formulation was assumed to follow a Weibull function and was estimated based on the concentration–time datasets after oral administration [18].
2.3 Model Evaluation
Model-based stochastic simulations were created for visual comparison with the observed concentration–time datasets of voriconazole in different CYP2C19 genotype groups. For clinical trials not reporting CYP2C19 genotype information, the population was assumed to be NM as this genotype is the most common 2C19 polymorphism prevalent in more than 64% of White, African American, Hispanic, and Ashkenazi populations [39]. To compare the variability of observed and simulated PK datasets, 68% population prediction intervals (approximately mean ± SD in case of assumed normal distribution) were plotted if the observed concentration–time datasets were reported as mean (± SD), while 95% population prediction intervals were described when all individual concentration–time datasets were available [40]. The visual criteria for a good model performance were that 95% population prediction intervals should cover the observed individual plasma concentration–time datasets, or that the observed aggregate plasma concentration–time datasets should be inside the 68% population prediction intervals. Predicted AUC, Cmax, and Ctrough values were compared with observed values via goodness-of-fit plots.
The quantitative evaluation criterion for a good model performance was that the ratios of predicted to observed AUC, Cmax, and Ctrough should be within 0.5- to 2.0-fold limits, as shown in Tables 1 and 2, and Table S4 in the ESM. As a quantitative summary of the predictive performance of the model, the geometric mean fold error (GMFE) was calculated using Eq. (1) [41].
where GMFE is the geometric mean fold error of all AUC, Cmax, or Ctrough predictions from the respective model, pred P is the predicted parameter (AUC, Cmax, or Ctrough), obs P is the observed parameter (AUC, Cmax, or Ctrough), and n is the number of studies.
2.4 Drug–Drug Interactions with Other CYP3A4 Substrates
Published PBPK models of the CYP3A4 probe substrates midazolam or alfentanil were integrated with the model of voriconazole to assess the inhibitory effects of voriconazole on CYP3A4 in vivo and to verify the inhibition model of voriconazole [41]. The DDI modeling performance was evaluated by both visual comparison of predicted versus observed probe substrate PK datasets, and by calculation of DDI AUC ratios and Cmax ratios according to Eqs. (2) and (3).
where AUC (or Cmax) treatment is the AUC (or Cmax) of victim drug with voriconazole co-treatment, and AUC (or Cmax) reference is the AUC (or Cmax) for victim drug administration alone.
2.5 Sensitivity Analysis
According to Eq. (4), the ratio of the relative change of AUC during a dosing interval (AUCτ) versus the relative alteration of the evaluated parameter was calculated at steady state after the standard therapeutic multiple dosings of voriconazole by oral administration. The sensitivity analysis was also conducted for the DDI between voriconazole and midazolam. Parameters selected for the sensitivity analysis fulfilled one of the following criteria [41]: (1) optimized; (2) related to optimized parameters; (3) a strong influence on calculation methods used in the model; and (4) significant impact in the model.
where S is the sensitivity of AUC to the evaluated parameter, ΔAUC is the change of AUC, AUC is the AUC with the initial value, Δp is the change of the assessed parameter value, and p is the parameter with the initial value. A sensitivity value of + 1.0 means that a 10% change in the examined parameter causes a 10% alteration of the predicted AUCτ.
In addition, we evaluated the uncertainty of inhibitory parameters KI and kinact by Monte Carlo simulations. First, 1000 pairs of KI and kinact values were randomly sampled based on the normal distribution of kinact of [point estimate and 95% confidence interval (CI)] 0.015 (0.011–0.019) min−1 and the log normal distribution of KI of 9.33 (2.56–34.0) µM; these 1000 pairs of parameters were then entered into the model to perform simulations of AUC and Cmax. Two scenarios were simulated. Scenario A was oral treatment of voriconazole 400 mg twice daily on the first day followed by 200 mg twice daily for 2 weeks, which was considered to be sufficient to achieve steady-state. AUCtlast−1_tlast and Cmax values of the last dosing interval were simulated. Scenario B was oral treatment of voriconazole 400 mg twice daily on the first day followed by 200 mg twice daily on the second day, and oral coadministration of midazolam 7.5 mg with the last dose of voriconazole. AUClast and Cmax values of voriconazole and midazolam for the last dose were simulated.
2.6 Virtual Population Characteristics
Based on the demographic characteristics from each clinical trial, virtual populations of 100 individuals were generated to quantitatively assess the variability of the predicted concentration–time datasets from the respective clinical trials. Information on age, body weight, body height and proportion of female participants was integrated into the software for each clinical trial. The default population variabilities for enzyme expression in PK-Sim® were used.
2.7 Model Applications
First, model-based simulations were performed according to the dosing regimens of the clinical trials in Table 1 to compare the predicted versus observed data, capturing the nonlinear PK of voriconazole including dose- and time-dependence. Second, different CYP2C19 genotype groups, i.e. RMs, NMs, intermediate metabolizers (IMs) and PMs were simulated respectively to depict the effect of genetic polymorphisms of CYP2C19 on the metabolism of voriconazole in Table 2. Based on the PBPK model, we then explored the performance of various maintenance doses in different CYP2C19 genotype groups (RMs, NMs, and IMs). Virtual populations of 1000 individuals were generated based on the summary demographic characteristics from all clinical trials. The simulated dosing regimens were 400 mg twice daily on the first day, followed by 100–400 mg twice daily on the following days for 2 weeks, which was considered to be sufficient to achieve steady-state. The trough plasma concentration sample was simulated to be taken prior to the last dose. The probability of target attainment and of reaching potentially toxic Ctrough values was calculated based on two different definitions of therapeutic ranges to reflect the heterogeneity of guidelines. Thus, a therapeutic target of Ctrough at least 1 or 2 mg/L and at most 5 or 6 mg/L was defined. Third, the time course of active CYP3A4 content in both the liver and small intestine during voriconazole treatment was simulated based on the most frequent oral therapeutic dosing regimen of voriconazole, i.e. 400 mg twice daily on the first day and then 200 mg twice daily on the following days. Fourth, by connecting the PBPK models of midazolam (or alfentanil) and voriconazole, DDI models between voriconazole and the victim drugs were set up (see Table 3).
3 Results
3.1 In Vitro Assays
The results of the half maximal inhibitory concentration (IC50) shift assays indicated that voriconazole caused TDI on CYP3A4, with a 16-fold difference in the absence and presence of nicotinamide adenine dinucleotide phosphate (NADPH) (see Table 4), supporting TDI to be introduced into the PBPK model. In contrast, inhibition of CYP2C19 was only within a two/threefold range of IC50 shift and was therefore considered as negligible during model development. The inactivation kinetic assay gave a KI of 9.33 (95% CI 2.56–34.0) μM and a kinact of 0.0428 (95% CI 0.0171–0.107) min−1 for CYP3A4, which were used for the parameterization in the PBPK model (see Table 5).
3.2 Model Development and Evaluation
3.2.1 Clinical Studies
Among all 93 concentration–time datasets of voriconazole from clinical trials, 21 were used for the model development and 72 were used for model evaluation (see Tables 1 and 2). The participants were all healthy volunteers, with an age range of 18–53 years and a body weight of 47–103 kg. CYP2C19 genotypes included 62 RMs (*1/*17, *17/*17), 101 NMs (*1/*1), 77 IMs (*1/*2, *1/*3, *2/*17, *2/*2/*17), and 65 PMs (*2/*2, *2/*3, *3/*3) (see Table 2). Administration protocols included both oral and intravenous routes, both single and multiple doses, and individual doses ranging from 1.5 to 6 mg/kg and from 50 to 400 mg.
3.2.2 Model Development
The input parameters describing the PBPK model of voriconazole are listed in Table 6. Vmax for CYP3A4 was originally fixed to 0.31 pmol/min/pmol according to the reported value by Damle et al. [31]. However, simulations resulted in a more than twofold overprediction for AUC for low doses of voriconazole. The reasons for overprediction of AUC were explored. Simultaneous and separate optimization of Vmax for CYP3A4 and CYP2C19 showed that the optimized value for CYP2C19 was approaching the value reported, while for CYP3A4, the optimized value was far higher than the reported value. A possible reason was that the reported value for CYP3A4 was obtained without consideration of TDI on CYP3A4, which might lead to underestimation of Vmax. Furthermore, the subjects in the clinical studies belonged to different CYP2C19 genotypes, which provided the possibility to optimize the Vmax of CYP3A4. Therefore, this parameter was optimized as 2.12 pmol/min/pmol based on the concentration–time datasets of CYP2C19 PMs with intravenous administration [18], assuming that only CYP3A4 mediated the metabolism of voriconazole in PMs due to the deficiency of CYP2C19. For other genotypes, both CYP2C19 and CYP3A4 contributed to the metabolism of voriconazole. The different CYP2C19 genotypes were integrated into the model for RMs, NMs, IMs or PMs, with the reference CYP2C19 expression values of 0.79, 0.76, 0.40 and 0.01 µmol/L, respectively [34]. Therefore, in the absence of evidence for another root cause of AUC overprediction, TDI of CYP3A4 by voriconazole was introduced into the model, assuming that it reflects MBI, with Eq. S4 in the ESM based on the in vitro inactivation kinetic parameter KI of 9.33 μM. When the in vitro kinact of 0.0428 min−1 served as the model input, the predicted concentration–time datasets of midazolam in DDI with co-treatment of voriconazole were overestimated. Therefore, kinact was finally optimized as 0.015 min−1 based on the concentration–time datasets with multiple intravenous dosing of voriconazole [36].
3.2.3 Model Evaluation
The predicted PK results for the respective clinical trials in comparison with the observed aggregate values are presented in Tables 1 and 2, together with administration protocols and subjects’ details. Prediction performance of the model was quantitatively evaluated by the ratios of predicted versus observed aggregate AUC and Cmax values, with calculated GMFEs being shown in Tables 1 and 2. Among the 28 test datasets for subjects with unspecified genotype, 71% of predicted/observed aggregate AUC ratios and all aggregate Cmax ratios were within the 0.5- to 2.0-fold limits (Table 1). Taking the genotype of CYP2C19 into consideration, from 44 test datasets, 89% of aggregate AUC ratios and all aggregate Cmax ratios were within 0.5- to 2.0-fold (Table 2). In addition, 85% of predicted/observed aggregate Ctrough ratios from clinical trials after multiple administration were within the 0.5- to 2.0-fold range (Table S4 in the ESM). The performance of the model was visualized by comparing predicted and observed concentration–time datasets, as shown in Figs. 3 and 4, and in Figs. S1, S2 and S4–7 in the ESM. The model-based simulations for multiple doses captured the dose- and time-dependent nonlinear PK of voriconazole well (Fig. 3, and Figs. S1, S4, and S7 in the ESM). Although the population predictions for low doses (i.e. 50 mg) reflected overestimation compared with the observed individual data, for the therapeutic dose of 400 mg the 95% prediction interval covered the variability of the observed individual data sufficiently (Fig. 4, and Fig. S5 in the ESM), indicating that simulations grouped by different CYP2C19 genotype were suitable to describe the effect of genetic polymorphisms of CYP2C19 on the metabolism of voriconazole. This was confirmed by the population predictions of observed aggregate concentration–time datasets for both single and multiple doses in different CYP2C19 genotype groups, despite an overprediction of exposure for multiple doses in PMs (Figs. S2 and S7 in the ESM). Furthermore, plotting predicted versus observed AUC, Cmax and Ctrough from all the clinical studies confirmed a good fit of the final PBPK model of voriconazole for most clinical trials (Fig. 5), while some overprediction of AUC values was present for low doses.
Prediction performance of the voriconazole PBPK model on aggregate plasma concentrations for multiple doses. Observed aggregate data reported in the literature are shown as a dot, triangle, square, cross, or crossed square [6, 36,37,38, 47,48,49,50,51,52,53,54,55,56,57,58,59,60]; population simulation medians are shown as lines; and the shaded areas illustrate the 68% population prediction intervals. Details of dosing regimens, study populations, and predicted versus observed PK parameters are summarized in Table 1. D day of treatment according to the numbering in the reference, qd once daily, bid twice daily, tid three times daily, IV intravenously, PO orally, Plasma conc voriconazole plasma concentration, PBPK physiologically based pharmacokinetic, PK pharmacokinetic
Prediction performance of the voriconazole PBPK model on individual plasma concentrations in different CYP2C19 genotype groups for a single dose. Observed individual data reported in the literature are shown as dots [18, 24, 62, 63]; population simulation medians are shown as lines; and the shaded areas illustrate the 95% population prediction intervals. Details of dosing regimens, study populations, and predicted versus observed PK parameters are summarized in Table 2. IV intravenously, PO oral, Plasma conc voriconazole plasma concentration, RM rapid metabolizers, NM normal metabolizers, IM intermediate metabolizers, PM poor metabolizers, Rengel Rengelshausen, PBPK physiologically based pharmacokinetic, PK pharmacokinetic, CYP cytochrome P450
Goodness-of-fit plot of the PBPK model of voriconazole. Predicted versus observed aggregate AUC (a), Cmax (b), and Ctrough (c) of voriconazole from all clinical studies. The identity line and 0.5- to 2.0-fold acceptance limits are shown as solid and dashed lines, respectively. Different colors represent different clinical trials. “*” after the study name shows that different genotype groups are displayed in the study. PBPK physiologically based pharmacokinetic, AUC area under the concentration–time curve, Cmax maximum concentration, Ctrough trough concentration
3.3 Sensitivity Analysis
A sensitivity analysis was performed based on the simulation of the therapeutic multiple oral dosing regimen (i.e. 400 mg twice daily on the first day and then 200 mg twice daily on the following days until reaching steady-state) to assess the impact of the parameters on the model. The voriconazole model was most sensitive to CYP2C19 kcat, Km, and fraction unbound values (all taken from the literature), with sensitivity values ranging from − 1.08 to 0.75 (Fig. S3a in the ESM). Analysis of the parameters for voriconazole/midazolam DDI models on the AUClast of midazolam showed that sensitivity was most pronounced for midazolam lipophilicity, CYP3A4 kinact and KI with the sensitivity values beyond − 1.0 or 1.0 (Fig. S3b in the ESM).
The assessment of the uncertainty of inhibitory parameters KI and kinact in scenario A showed that simulated AUCtlast−1_tlast of voriconazole was (point estimate and 90% CI) 12.6 (7.77–16.4) mg/L*h and Cmax was 2.61 (2.02–3.01) mg/L, corresponding to a 90% CI of 61.6–130% of the point estimate for AUCtlast−1_tlast and 77.4–115% for Cmax. The simulation of scenario B resulted in voriconazole AUClast values of 14.1 (7.67–22.3) mg/L*h and Cmax values of 2.46 (1.86–3.05) mg/L; and midazolam AUClast values of 0.753 (0.227–1.84) mg/L*h and Cmax values of 0.121 (0.0751–0.149) mg/L. This corresponded to relative 90% CIs for voriconazole AUClast of 54.4–158% and Cmax of 75.6–124%; and for midazolam AUClast of 30.3–244% and Cmax of 62.1–123% of the respective point estimates.
3.4 Model Application
3.4.1 Suitable Maintenance Doses in CYP2C19 Genotype Groups
A separate simulation of specific CYP2C19 genotype groups could reasonably describe both observed individual and aggregate concentration–time datasets for either a single dose or multiple doses, as assessed by the respective criteria (Table 2 and Fig. 3, and Figs. S2, S5, and S7 in the ESM). Therefore, model-based simulations were carried out to explore the performance of voriconazole maintenance doses for different CYP2C19 genotypes (Fig. 8). The standard dosage (oral 400 mg twice daily on the first day and 200 mg twice daily for the following days) was confirmed to be appropriate for IMs, while for RMs and NMs, the 200 mg maintenance dose provided an insufficient exposure, with a probability of target attainment of < 30%. The results of model-based simulations showed that doubling the maintenance dose for RMs and NMs could increase the probability of target attainment twofold while maintaining a probability of reaching toxic concentrations below 20%. The less reliable prediction for multiple doses in PMs precludes the suggestion of an appropriate maintenance dose regimen in PMs, although it clearly shows that the 200 mg twice daily dose is too high.
3.4.2 Inhibition of CYP3A4 by Voriconazole
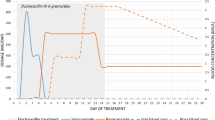
The time courses of CYP3A4 activity in both the liver and small intestine were assessed during long-term voriconazole treatment. The maximum inhibition was reached at 51.2 h in the liver and 52.5 h in the small intestine (Fig. 6), resulting from the combination of the physiological CYP3A4 turnover and TDI (in our model, MBI) of CYP3A4 (Eq. S4 in the ESM). The CYP3A activity was predicted to recover 90% of its baseline 5 days after the last voriconazole dose.
Effect of therapeutic multiple oral dosings of voriconazole on hepatic and small intestinal CYP3A activity. Predicted change of relative hepatic (green line) and small intestinal (red line) CYP3A activity over time after therapeutic multiple oral dosings of voriconazole. The blue line represents voriconazole plasma concentration, and arrows indicate dosing events of a standard therapeutic dosing schedule for oral voriconazole. CYP cytochrome P450
3.4.3 Drug–Drug Interaction Modeling
The CYP3A4 inhibition model of voriconazole was further applied to the DDI between CYP3A4 probe substrates as victims (midazolam and alfentanil) and voriconazole as the perpetrator. Figure 7, and Fig. S8 in the ESM, demonstrate the good performance of DDI PBPK models for voriconazole and the two probe substrates. The observed AUC change of substrates during co-treatment with voriconazole was inside the 90% CI of the predicted AUC change. For alfentanil, the predicted/observed DDI AUC ratio of alfentanil was 0.86, indicating that this inhibition model was appropriate (Table 3). The inhibition model was further confirmed to be suitable by the predicted/observed midazolam DDI AUC ratios of 1.09 and 0.76, respectively, for intravenous and oral administration of midazolam (Table 3).
Prediction performance of voriconazole PBPK model in DDIs with CYP3A4 probe substrates. The voriconazole model integrated with the models of CYP3A4 probe substrates predicted inhibitory effects of voriconazole on CYP3A4 in vivo. Population predictions of a alfentanil or b, c midazolam plasma concentration–time datasets, with and without voriconazole treatment, were compared with observed data shown as green triangles (control), red dots (voriconazole coadministration) or symbols ± SD [23, 66]. Population simulation medians are shown as green lines (control) or red lines (voriconazole coadministration), and the shaded areas illustrate the respective a 68% and b, c 95% population prediction intervals. Details of dosing regimens, study populations, and predicted and observed DDI AUC ratios and Cmax ratios are summarized in Table 3. IV intravenously, PO orally, PBPK physiologically based pharmacokinetic, DDI drug–drug interactions, CYP cytochrome P450, SD standard deviation, AUC area under the plasma concentration–time curve, Cmax maximum concentration
4 Discussion
A whole-body PBPK model of voriconazole integrating TDI of CYP3A4 has been successfully developed. Model-based simulations of voriconazole plasma concentrations were in good agreement with observations from clinical studies with both intravenous and oral administration of a wide range of single and multiple doses. The model was also appropriate to predict voriconazole plasma concentrations for individual CYP2C19 genotype groups and the extent of DDIs with the CPY3A4 probe substrates midazolam and alfentanil caused by voriconazole.
Several lines of evidence supported that the incorporation of TDI should be considered to accurately describe the PK of voriconazole. First, Mikus and colleagues proposed that ‘autoinhibition’ of CYP3A was the key to explaining the observed dose nonlinearity of voriconazole elimination after administration of 50 and 400 mg in healthy volunteers [15, 24]. Second, time-dependent disproportionately increasing exposure of voriconazole was found in vivo after multiple doses, e.g. AUC for multiple intravenous administration (3 mg kg−1 over 1 h once on the first day and twice daily on the following days) on the fifth day of treatment was more than twofold higher than the predicted value based on the results for the first dose under the assumption of dose-linearity, and continued to increase until the 12th-day doses [36]. Third, both Friberg et al. and Kim et al. integrated ‘time-dependent inhibition’ or ‘autoinhibition’ in their models to describe the respective processes regarding enzyme inhibition by voriconazole in vivo [25, 26]. Fourth, our in vitro assays clearly showed a pronounced IC50 shift from 48.7 to 3 µM, verifying TDI of CYP3A4 by voriconazole (Table 4). Indeed, incorporation of TDI (assuming MBI) into the PBPK model turned out to be essential to predict the dose- and time-dependent PK nonlinearity of voriconazole.
Beyond TDI, reversible inhibition of CYP3A4 and CYP2C19 by voriconazole was also explored. Our in vitro assay resulted in a competitive inhibition of CYP3A4 Ki of 0.47 (95% CI 0.344–0.636) µM, which is in agreement with the results from other studies, e.g. competitive inhibition (Ki = 0.66 µM) and noncompetitive inhibition (Ki = 2.97 µM) in one study [21], and solely competitive inhibition (Ki = 0.15 µM) in another study [22]. However, in vivo evaluation of DDIs between voriconazole and midazolam indicated that assumption of a simple competitive inhibition only was explicitly not sufficient in vivo [42]. A TDI model of CYP3A was discussed in the previous research but was not incorporated due to a lack of in vitro data to support it. At that time, a hypothetical extra effect compartment was introduced to describe a time delay [42]. Thus, we conducted an in vitro assay to explore TDI of voriconazole on CYP3A4 to fully understand the metabolism of voriconazole.
Furthermore, our in vitro assay showed competitive inhibition of voriconazole on CYP2C19 with Ki values of 1.08 (95% CI 0.815–1.43) µM and 1.26 (95% CI 0.839–1.82) µM using omeprazole and mephenytoin as substrates, respectively (see Table 4), which could provide some evidence for DDIs between voriconazole and CYP2C19 probe substrates (e.g. omeprazole and mephenytoin). In vivo, voriconazole was reported to increase the Cmax and AUCτ of omeprazole by 116% and 280% [43], respectively. However, detailed in vivo data were not available, which limited the evaluation of the PBPK DDI models between voriconazole and CYP2C19 substrates, which is one of the limitations of our PBPK model.
Beyond the effects of the parent drug, the inhibition of voriconazole N-oxide on CYP3A4 and CYP2C19 was also investigated. Although voriconazole N-oxide exhibited reversible inhibition on both enzymes, the effects were weaker with Ki 0.894 (95% CI 0.650–1.22) and 9.00 (95% CI 6.94–11.7) µM, respectively (see Table 4). Additionally, at therapeutic voriconazole doses, plasma concentrations of voriconazole N-oxide typically reach only about one-third compared with that of its parent drug [17]. Thus, the inhibition by voriconazole N-oxide would be much less than that of the parent drug and was considered negligible during PBPK model development.
The advantages of the PBPK model approach presented here becomes evident when compared with an empirical population PK model. PBPK models can provide a more precise mechanistic picture of inhibition processes. Based on the developed PBPK model, it was feasible to describe the time course of inhibition of CYP3A4 during and after voriconazole treatment by taking into account the dynamic nature of the inhibition process, with a clear differentiation between liver and small intestinal enzyme activity (Fig. 6). Furthermore, this PBPK model could be applied to predict the effect of voriconazole dosing schemes on other CYP3A4 substrate drugs, and to thus manage respective clinical DDIs. This was verified by the observation that the prediction of DDIs was mostly appropriate for oral and intravenous midazolam, as well as for alfentanil (Fig. 7, and Fig. S8 in the ESM), both being established CYP3A4 probe substrates [44].
For a thorough understanding of voriconazole PK, CYP2C19 genotype groups were another important factor during model development since the wide interindividual variability mainly results from differences in enzyme activity between CYP2C19 genotypes. Therefore, suitable maintenance doses for the CYP2C19 genotype groups (RMs, NMs, and IMs) were suggested based on simulations. For PMs, the search for a dose to provide an appropriate exposure was less reliable due to the limited performance of the model for multiple doses in this genotype group. With TDI on CYP3A4 activity and deficiency of CYP2C19, voriconazole would accumulate in PMs and might reach extremely high concentrations after multiple administrations. However, the observations from one study showed that the increase of voriconazole concentrations in PMs after multiple doses was less than predicted (Fig. S2f in the ESM) [19], indicating that other elimination pathways may compensate and thus attenuate drug accumulation in the body. However, for PMs, the experimental data to quantitatively describe voriconazole PK in individuals were sparse, limiting the integration of more complex pathways.
Although the presented model performed well with respect to both single and multiple doses and in most CYP2C19 genotype groups (RMs, NMs, and IMs), it has several limitations. First is the assumption that only CYP3A4 and CYP2C19 mediate primary metabolism and elimination of voriconazole. This assumption may result in overestimation of the role of CYP3A4 and CYP2C19 activity; however, the consequence of ignoring FMO and CYP2C9 should be acceptable in most CYP2C19 genotypes (RMs, NMs, and IMs). Km values for FMO1 and FMO3 are in the millimolar range (about 3 mM) [14], which is far beyond the concentrations reached in vivo. A contribution of CYP2C9 was identified in only one paper [13] with a small Vmax value, which was not confirmed in other in vitro assays [13, 45]. Renal excretion of unchanged voriconazole is < 2%, and primary metabolism by glucuronidation is also negligible [17]. Thus, it is reasonable to simplify the primary metabolism of voriconazole as depending on CYP3A4 and CYP2C19 only. In addition, the fact that our model was able to properly describe most published data supports the pivotal role of CYP3A4 and CYP2C19 for overall voriconazole elimination. Another limitation is that the minor inhibitory effect of voriconazole N-oxide observed in vitro, as well as possible effects of other voriconazole metabolites, were not taken into account. In addition, we did not attempt to simultaneously describe the concentration–time datasets of voriconazole N-oxide and other metabolites (hydroxy-fluoropyrimidine voriconazole and dihydroxy-fluoropyrimidine voriconazole) reported in a few published studies to limit the complexity of the model and to limit the number of assumptions required. The third limitation was that during model development, datasets with low voriconazole doses, e.g. 50 mg, were not successfully integrated into the model. When extrapolating the model predictions to low dosages, the simulation showed some overprediction of voriconazole concentrations; however, such low doses are not clinically relevant. Fourth, based on the datasets of healthy volunteers, the model-based simulations provided suggestions for an appropriate dosage for CYP2C19 genotype subgroup (see Fig. 8). However, the applicability of modeling results for patients needs to be confirmed in future studies. Currently, therapeutic drug monitoring for voriconazole would be preferred for all patient subgroups to guarantee proper voriconazole concentrations in each patient. Fifth, while an all-embracing assessment of all uncertainties of input parameters on various potential model outcomes was not feasible, we performed an assessment of the uncertainty of the key parameters. i.e. KI and kinact. While the 90% CI of the resulting distribution for the exposure of voriconazole itself was within the 0.5- to 2-fold range of its median in the model, the respective simulated 90% CI for midazolam exposure slightly exceeded a twofold deviation from the median. However, in light of the observed high variability in exposure changes of midazolam when coadministered with voriconazole, we concluded that the uncertainty of the inhibitory parameters is acceptable in our model, in particular given the fact that a potential covariance of KI and kinact was neglected for parameter sampling. On the other hand, the need to optimize the experimentally obtained kinact based on clinical data may also reflect the limitations of our in vitro experiments to quantitatively predict enzyme inhibition in vivo.
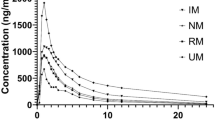
Probability of target attainment for therapeutic and toxic Ctrough in different CYP2C19 genotype groups for long-term dosing. The simulated dosing regimens were 400 mg bid on the first day, followed by 100–400 mg bid on the following days for 2 weeks. The final trough plasma concentration sample was simulated to be taken prior to the last dose. Red and green lines represent the probability of therapeutic target attainment based on Ctrough above 1 mg/L and above 2 mg/L, respectively. Blue and purple lines show the probability of toxicity target attainment based on Ctrough above 5 mg/L and above 6 mg/L, respectively. Black lines show the optimal dose for each genotype group. IM intermediate metabolizers, NM normal metabolizers, RM rapid metabolizers, Ctrough trough concentration, CYP cytochrome P450, bid twice daily
Although the current model successfully described the complex metabolism of voriconazole, we suggest to further verify the model by additional in vitro studies (e.g. elucidating the exact mechanism of TDI on CYP3A4) and clinical studies (e.g. studies quantifying the metabolites of voriconazole, i.e. voriconazole N-oxide, hydroxy-fluoropyrimidine voriconazole, and dihydroxy-fluoropyrimidine voriconazole in plasma/urine/feces; studies in PMs with low multiple doses; and DDI studies between CYP3A4 substrates and voriconazole, including quantification of its metabolites and different routes of administration of both substrates and voriconazole).
5 Conclusions
TDI of CYP3A4 by voriconazole is an important PK characteristic of the drug and needs to be taken into account, along with the CYP2C19 genotype, to properly predict the exposure of voriconazole. By incorporating these elements, a PBPK model of voriconazole was developed that could accurately capture the time- and dose-dependent alterations of voriconazole PK, as well as DDIs caused by voriconazole inhibitory effects on CYP3A4. This model could support individual dose optimization of voriconazole, as well as DDI risk management. It will be provided as a public tool in the OSP repository (http://www.open-systems-pharmacology.org/) to assess the DDI potential of investigational drugs, support the design of clinical trials, or to expand the model for predictions in special populations.
References
US FDA. Pfizer Label: voriconazole for injection, tablets, oral suspension: LAB-0271-12. US FDA; 2005.
Herbrecht R, Denning DW, Patterson TF, Bennett JE, Greene RE, Oestmann J-W, et al. Voriconazole versus amphotericin B for primary therapy of invasive aspergillosis. N Engl J Med. 2002;347:408–15.
Misch EA, Safdar N. Updated guidelines for the diagnosis and management of aspergillosis. J Thorac Dis. 2016;8:E1771–6.
Ullmann AJ, Aguado JM, Arikan-Akdagli S, Denning DW, Groll AH, Lagrou K, et al. Diagnosis and management of Aspergillus diseases: executive summary of the 2017 ESCMID-ECMM-ERS guideline. Clin Microbiol Infect. 2018;24:e1–38.
Theuretzbacher U, Ihle F, Derendorf H. Pharmacokinetic/pharmacodynamic profile of voriconazole. Clin Pharmacokinet. 2006;45:649–63.
Purkins L, Wood N, Ghahramani P, Greenhalgh K, Allen MJ, Kleinermans D. Pharmacokinetics and safety of voriconazole following intravenous- to oral-dose escalation regimens. Antimicrob Agents Chemother. 2002;46:2546–53.
Owusu Obeng A, Egelund EF, Alsultan A, Peloquin CA, Johnson JA. CYP2C19 polymorphisms and therapeutic drug monitoring of voriconazole: are we ready for clinical implementation of pharmacogenomics? Pharmacotherapy. 2014;34:703–18.
Pascual A, Calandra T, Bolay S, Buclin T, Bille J, Marchetti O. Voriconazole therapeutic drug monitoring in patients with invasive mycoses improves efficacy and safety outcomes. Clin Infect Dis. 2008;46:201–11.
Jin H, Wang T, Falcione BA, Olsen KM, Chen K, Tang H, et al. Trough concentration of voriconazole and its relationship with efficacy and safety: a systematic review and meta-analysis. J Antimicrob Chemother. 2016;71:1772–85.
Hamada Y, Tokimatsu I, Mikamo H, Kimura M, Seki M, Takakura S, et al. Practice guidelines for therapeutic drug monitoring of voriconazole: a consensus review of the Japanese Society of Chemotherapy and the Japanese Society of Therapeutic Drug Monitoring. J Infect Chemother. 2013;19:381–92.
Ashbee HR, Barnes RA, Johnson EM, Richardson MD, Gorton R, Hope WW. Therapeutic drug monitoring (TDM) of antifungal agents: guidelines from the British Society for Medical Mycology. J Antimicrob Chemother. 2014;69:1162–76.
Chen K, Zhang X, Ke X, Du G, Yang K, Zhai S. Individualized medication of voriconazole: a practice guideline of the division of therapeutic drug monitoring, Chinese Pharmacological Society. Ther Drug Monit. 2018;40:663–74.
Hyland R, Jones BC, Smith DA. Identification of the cytochrome P450 enzymes involved in the N-oxidation of voriconazole. Drug Metab Dispos. 2003;31:540–7.
Yanni SB, Annaert PP, Augustijns P, Bridges A, Gao Y, Benjamin DK, et al. Role of flavin-containing monooxygenase in oxidative metabolism of voriconazole by human liver microsomes. Drug Metab Dispos. 2008;36:1119–25.
Hohmann N, Kreuter R, Blank A, Weiss J, Burhenne J, Haefeli WE, et al. Autoinhibitory properties of the parent but not of the N-oxide metabolite contribute to infusion rate-dependent voriconazole pharmacokinetics. Br J Clin Pharmacol. 2017;83:1954–65.
Roffey SJ, Cole S, Comby P, Gibson D, Jezequel SG, Nedderman ANR, et al. The disposition of voriconazole in mouse, rat, rabbit, guinea pig, dog, and human. Drug Metab Dispos. 2003;31:731–41.
Geist MJP, Egerer G, Burhenne J, Riedel K-D, Weiss J, Mikus G. Steady-state pharmacokinetics and metabolism of voriconazole in patients. J Antimicrob Chemother. 2013;68:2592–9.
Scholz I, Oberwittler H, Riedel K-D, Burhenne J, Weiss J, Haefeli WE, et al. Pharmacokinetics, metabolism and bioavailability of the triazole antifungal agent voriconazole in relation to CYP2C19 genotype. Br J Clin Pharmacol. 2009;68:906–15.
Lee S, Kim B-H, Nam W-S, Yoon SH, Cho J-Y, Shin S-G, et al. Effect of CYP2C19 polymorphism on the pharmacokinetics of voriconazole after single and multiple doses in healthy volunteers. J Clin Pharmacol. 2012;52:195–203.
Weiss J, ten Hoevel MM, Burhenne J, Walter-Sack I, Hoffmann MM, Rengelshausen J, et al. CYP2C19 genotype is a major factor contributing to the highly variable pharmacokinetics of voriconazole. J Clin Pharmacol. 2009;49:196–204.
Jeong S, Nguyen PD, Desta Z. Comprehensive in vitro analysis of voriconazole inhibition of eight cytochrome P450 (CYP) enzymes: major effect on CYPs 2B6, 2C9, 2C19, and 3A. Antimicrob Agents Chemother. 2009;53:541–51.
Yamazaki H, Nakamoto M, Shimizu M, Murayama N, Niwa T. Potential impact of cytochrome P450 3A5 in human liver on drug interactions with triazoles. Br J Clin Pharmacol. 2010;69:593–7.
Saari TI, Laine K, Leino K, Valtonen M, Neuvonen PJ, Olkkola KT. Effect of voriconazole on the pharmacokinetics and pharmacodynamics of intravenous and oral midazolam. Clin Pharmacol Ther. 2006;79:362–70.
Hohmann N, Kocheise F, Carls A, Burhenne J, Weiss J, Haefeli WE, et al. Dose-dependent bioavailability and CYP3A Inhibition contribute to non-linear pharmacokinetics of voriconazole. Clin Pharmacokinet. 2016;55:1535–45.
Friberg LE, Ravva P, Karlsson MO, Liu P. Integrated population pharmacokinetic analysis of voriconazole in children, adolescents, and adults. Antimicrob Agents Chemother. 2012;56:3032–42.
Kim Y, Rhee S-J, Park WB, Yu K-S, Jang I-J, Lee S. A personalized CYP2C19 phenotype-guided dosing regimen of voriconazole using a population pharmacokinetic analysis. J Clin Med. 2019;8:227–41.
Li X, Frechen S, Moj D, Taubert M, Hsin C, Mikus G, et al. A physiologically-based pharmacokinetic model of voriconazole (abstract no. 8995). Population Approach Group in Europe; 2019. ISSN 1871-6032.
Davies B, Morris T. Physiological parameters in laboratory animals and humans. Pharm Res. 1993;10:1093–5.
Edginton AN, Schmitt W, Willmann S. Development and evaluation of a generic physiologically based pharmacokinetic model for children. Clin Pharmacokinet. 2006;45:1013–34.
Mordenti J. Man versus beast: pharmacokinetic scaling in mammals. J Pharm Sci. 1986;75:1028–40.
Damle B, Varma MV, Wood N. Pharmacokinetics of voriconazole administered concomitantly with fluconazole and population-based simulation for sequential use. Antimicrob Agents Chemother. 2011;55:5172–7.
Meyer M, Schneckener S, Ludewig B, Kuepfer L, Lippert J, Weinstein S. Using expression data for quantification of active processes in physiologically based pharmacokinetic modeling. Drug Metab Dispos. 2012;40:892–901.
Rodrigues AD. Integrated cytochrome P450 reaction phenotyping: attempting to bridge the gap between cDNA-expressed cytochromes P450 and native human liver microsomes. Biochem Pharmacol. 1999;57:465–80.
Shirasaka Y, Chaudhry AS, McDonald M, Prasad B, Wong T, Calamia JC, et al. Interindividual variability of CYP2C19-catalyzed drug metabolism due to differences in gene diplotypes and cytochrome P450 oxidoreductase content. Pharmacogenomics J. 2016;16:375–87.
Michaelis L, Menten ML, Johnson KA, Goody RS. The original Michaelis constant: translation of the 1913 Michaelis–Menten paper. Biochemistry. 2011;50:8264–9.
Purkins L, Wood N, Greenhalgh K, Eve MD, Oliver SD, Nichols D. The pharmacokinetics and safety of intravenous voriconazole-a novel wide-spectrum antifungal agent. Br J Clin Pharmacol. 2003;56:2–9.
Purkins L, Wood N, Kleinermans D, Love ER. No clinically significant pharmacokinetic interactions between voriconazole and indinavir in healthy volunteers. Br J Clin Pharmacol. 2003;56(Suppl 1):62–8.
Purkins L, Wood N, Kleinermans D, Greenhalgh K, Nichols D. Effect of food on the pharmacokinetics of multiple-dose oral voriconazole. Br J Clin Pharmacol. 2003;56:17–23.
Strom CM, Goos D, Crossley B, Zhang K, Buller-Burkle A, Jarvis M, et al. Testing for variants in CYP2C19: population frequencies and testing experience in a clinical laboratory. Genet Med. 2012;14:95–100.
European Medicines Agency. Guideline on the reporting of physiologically based pharmacokinetic (PBPK) modelling and simulation. EMA/CHMP/458101/2016. EMA; 2018.
Hanke N, Frechen S, Moj D, Britz H, Eissing T, Wendl T, et al. PBPK models for CYP3A4 and P-gp DDI prediction: a modeling network of rifampicin, itraconazole, clarithromycin, midazolam, alfentanil, and digoxin. CPT Pharmacomet Syst Pharmacol. 2018;7:647–59.
Frechen S, Junge L, Saari TI, Suleiman AA, Rokitta D, Neuvonen PJ, et al. A semiphysiological population pharmacokinetic model for dynamic inhibition of liver and gut wall cytochrome P450 3A by voriconazole. Clin Pharmacokinet. 2013;52:763–81.
Donnelly JP, De Pauw BE. Voriconazole—a new therapeutic agent with an extended spectrum of antifungal activity. Clin Microbiol Infect. 2004;10:107–17.
Fuhr U, Hsin C, Li X, Jabrane W, Sörgel F. Assessment of pharmacokinetic drug-drug interactions in humans: in vivo probe substrates for drug metabolism and drug transport revisited. Annu Rev Pharmacol Toxicol. 2019;59:507–36.
Schulz J, Kluwe F, Mikus G, Michelet R, Kloft C. Novel insights into the complex pharmacokinetics of voriconazole: a review of its metabolism. Drug Metab Rev. 2019;51:247–65.
Chung H, Lee H, Han H, An H, Lim KS, Lee Y, et al. A pharmacokinetic comparison of two voriconazole formulations and the effect of CYP2C19 polymorphism on their pharmacokinetic profiles. Drug Des Dev Ther. 2015;9:2609–16.
Purkins L, Wood N, Greenhalgh K, Allen MJ, Oliver SD. Voriconazole, a novel wide-spectrum triazole: oral pharmacokinetics and safety. Br J Clin Pharmacol. 2003;56(Suppl 1):10–6.
Wood N, Tan K, Purkins L, Layton G, Hamlin J, Kleinermans D, et al. Effect of omeprazole on the steady-state pharmacokinetics of voriconazole. Br J Clin Pharmacol. 2003;56(Suppl 1):56–61.
Keirns J, Sawamoto T, Holum M, Buell D, Wisemandle W, Alak A. Steady-state pharmacokinetics of micafungin and voriconazole after separate and concomitant dosing in healthy adults. Antimicrob Agents Chemother. 2007;51:787–90.
Liu P, Foster G, Gandelman K, LaBadie RR, Allison MJ, Gutierrez MJ, et al. Steady-state pharmacokinetic and safety profiles of voriconazole and ritonavir in healthy male subjects. Antimicrob Agents Chemother. 2007;51:3617–26.
Purkins L, Wood N, Ghahramani P, Kleinermans D, Layton G, Nichols D. No clinically significant effect of erythromycin or azithromycin on the pharmacokinetics of voriconazole in healthy male volunteers. Br J Clin Pharmacol. 2003;56:30–6.
Purkins L, Wood N, Kleinermans D, Nichols D. Histamine H2-receptor antagonists have no clinically significant effect on the steady-state pharmacokinetics of voriconazole. Br J Clin Pharmacol. 2003;56(Suppl 1):51–5.
Purkins L, Wood N, Ghahramani P, Love ER, Eve MD, Fielding A. Coadministration of voriconazole and phenytoin: pharmacokinetic interaction, safety, and toleration. Br J Clin Pharmacol. 2003;56(Suppl 1):37–44.
Marshall WL, McCrea JB, Macha S, Menzel K, Liu F, van Schanke A, et al. Pharmacokinetics and tolerability of letermovir coadministered with azole antifungals (posaconazole or voriconazole) in healthy subjects. J Clin Pharmacol. 2018;58:897–904.
Liu P, Foster G, LaBadie RR, Gutierrez MJ, Sharma A. Pharmacokinetic interaction between voriconazole and efavirenz at steady state in healthy male subjects. J Clin Pharmacol. 2008;48:73–84.
Andrews E, Damle BD, Fang A, Foster G, Crownover P, LaBadie R, et al. Pharmacokinetics and tolerability of voriconazole and a combination oral contraceptive co-administered in healthy female subjects. Br J Clin Pharmacol. 2008;65:531–9.
Damle B, LaBadie R, Crownover P, Glue P. Pharmacokinetic interactions of efavirenz and voriconazole in healthy volunteers. Br J Clin Pharmacol. 2008;65:523–30.
Dodds Ashley ES, Zaas AK, Fang AF, Damle B, Perfect JR. Comparative pharmacokinetics of voriconazole administered orally as either crushed or whole tablets. Antimicrob Agents Chemother. 2007;51:877–80.
Kakuda TN, Van Solingen-Ristea R, Aharchi F, De Smedt G, Witek J, Nijs S, et al. Pharmacokinetics and short-term safety of etravirine in combination with fluconazole or voriconazole in HIV-negative volunteers. J Clin Pharmacol. 2013;53:41–50.
Dowell JA, Schranz J, Baruch A, Foster G. Safety and pharmacokinetics of coadministered voriconazole and anidulafungin. J Clin Pharmacol. 2005;45:1373–82.
Wang G, Lei H, Li Z, Tan Z, Guo D, Fan L, et al. The CYP2C19 ultra-rapid metabolizer genotype influences the pharmacokinetics of voriconazole in healthy male volunteers. Eur J Clin Pharmacol. 2009;65:281–5.
Mikus G, Schöwel V, Drzewinska M, Rengelshausen J, Ding R, Riedel KD, et al. Potent cytochrome P450 2C19 genotype-related interaction between voriconazole and the cytochrome P450 3A4 inhibitor ritonavir. Clin Pharmacol Ther. 2006;80:126–35.
Rengelshausen J, Banfield M, Riedel K, Burhenne J, Weiss J, Thomsen T, et al. Opposite effects of short-term and long-term St John’s wort intake on voriconazole pharmacokinetics. Clin Pharmacol Ther. 2005;78:25–33.
Lei H-P, Wang G, Wang L-S, Ou-yang D, Chen H, Li Q, et al. Lack of effect of ginkgo biloba on voriconazole pharmacokinetics in Chinese volunteers identified as CYP2C19 poor and extensive metabolizers. Ann Pharmacother. 2009;43:726–31.
Zhu L, Brüggemann RJ, Uy J, Colbers A, Hruska MW, Chung E, et al. CYP2C19 genotype-dependent pharmacokinetic drug interaction between voriconazole and ritonavir-boosted atazanavir in healthy subjects. J Clin Pharmacol. 2017;57:235–46.
Saari TI, Laine K, Leino K, Valtonen M, Neuvonen PJ, Olkkola KT. Voriconazole, but not terbinafine, markedly reduces alfentanil clearance and prolongs its half-life. Clin Pharmacol Ther. 2006;80:502–8.
Zane NR, Thakker DR. A physiologically based pharmacokinetic model for voriconazole disposition predicts intestinal first-pass metabolism in children. Clin Pharmacokinet. 2014;53(12):1171–82.
Qi F, Zhu L, Li N, Ge T, Xu G, Liao S. Influence of different proton pump inhibitors on the pharmacokinetics of voriconazole. Int J Antimicrob Agents. 2017;49(4):403–9.
Pfizer. Vfend: voriconazole for IV infusion safety data sheet. 2018. https://pfe-pfizercom-prod.s3.amazonaws.com/products/material_safety_data/voriconazole_IV_infusion_22-mar-2018.pdf.
Pfizer Canada Inc. VFEND® voriconazole tablets 50 mg and 200 mg for injection 200 mg / vial (10 mg/mL when reconstituted) powder for oral suspension 3 g / bottle (40 mg/mL when reconstituted) submission control No: 196793. Kirkland, Quebec; 2016. https://www.pfizer.ca/sites/default/files/201710/VFEND_PM_E_196793_23Sept2016.pdf.
Scientific Discussion-VFEND Procedure No. EMEA/H/C/387/X/09. [London]: London; 2004. https://www.ema.europa.eu/en/documents/scientific-discussion-variation/vfend-h-c-387-x-0009-epar-scientific-discussion-extension_en.pdf.
Murayama N, Imai N, Nakane T, Shimizu M, Yamazaki H. Roles of CYP3A4 and CYP2C19 in methyl hydroxylated and N-oxidized metabolite formation from voriconazole, a new anti-fungal agent, in human liver microsomes. Biochem Pharmacol. 2007;73:2020–6.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
XL obtained financial support provided by the China Scholarship Council during the study and during manuscript preparation (No. 201406920024). TIS obtained a governmental research grant (#13821) from the Hospital District of South-West Finland to support his work.
Conflict of interest
Sebastian Frechen is an employee and potential shareholder of Bayer AG, Leverkusen, Germany. Xia Li, Daniel Moj, Thorsten Lehr, Max Taubert, Chih-hsuan Hsin, Gerd Mikus, Pertti J. Neuvonen, Klaus T. Olkkola, Teijo I. Saari, and Uwe Fuhr have no conflicts of interest to declare.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, X., Frechen, S., Moj, D. et al. A Physiologically Based Pharmacokinetic Model of Voriconazole Integrating Time-Dependent Inhibition of CYP3A4, Genetic Polymorphisms of CYP2C19 and Predictions of Drug–Drug Interactions. Clin Pharmacokinet 59, 781–808 (2020). https://doi.org/10.1007/s40262-019-00856-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40262-019-00856-z