Abstract
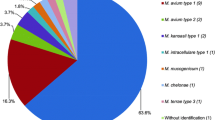
Out of 12 nontuberculous mycobacteria (NTM) species, 5 were identified as Mycobacterium tuberculosis (MTB) by GeneXpert at a bacterial load of 106. Notably, two species, including M. abscessus and M. smegmatis, were flagged as RIF-resistant MTB due to the high C(t) value of Probe E. In conclusion, our data have demonstrated that the misdiagnosis of MTB by GeneXpert assay is observed in five NTM species at a high bacterial load.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The GeneXpert MTB/RIF assay, developed by Cepheid Diagnostics (Sunnyvale, CA, USA), is a novel, semiautomatic nucleic-acid amplification test (NAAT) for simultaneously detecting Mycobacterium tuberculosis (MTB) and resistance to rifampin (RIF) from clinical specimens [1, 2]. The cartridge-based system integrates DNA extraction, rpoB gene amplification and sequence analysis in a single cartridge, thus requiring minimal laboratory measures and reducing the potential contamination risk [3, 4]. In addition, clinical evaluations revealed that this assay exhibited high sensitivity and specificity for both smear-positive and smear-negative clinical specimens [2]. Therefore, World Health Organization (WHO) endorsed the GeneXpert MTB/RIF assay to directly screen sputum samples for the rapid diagnosis of tuberculosis (TB), as well as RIF resistance [5].
The rpoB gene, encoding the β-subunit of RNA polymerase, is the chromosomal target of GeneXpert assay for detecting MTB and its susceptibility to RIF [6]. Five molecular beacons with different colors have been designed for hybridizing to different target segments within rpoB core region. Several previous studies have demonstrated that GeneXpert exhibits a satisfactory performance to distinguish MTB from nontuberculous mycobacteria (NTM) at a low bacterial load [3, 4, 7, 8], while the data regarding the efficiency of GeneXpert for differentiating NTM from MTB under different dilution was limited. Hence, the study aimed to investigate whether GeneXpert assay could yield true-negative results on serial dilutions of NTM suspension, especially at a high bacterial load.
Materials and methods
Bacterial strains
A total of 12 most common NTM species were obtained from National Tuberculosis Reference Laboratory, including M. marinum, M. abscessus, M. smegmatis, M. phlei, M. aurum, M. avium, M. intracellulare, M. kansasii, M. chelonae, M. scrofulaceum, M. gordonae and M. fortuitum, which were ordered from the American Type Culture Collection (ATCC), the global biological materials resource and standards organization. Prior to GeneXpert analysis, all the strains were identified by multi-locus DNA sequencing as previously reported [9]. Then the strains were subcultured on the Löwenstein-Jensen (L-J) slants for 1–4 weeks at 37 °C according to the growth rates of different NTM species.
Preparation of inoculum and GeneXpert analysis
The freshly grown bacterial colonies were harvested from the L-J slants, and transferred to a screw-cap tube with glass beads and 1 mL normal saline. After vigorous agitation for 30 s, the supernatant suspension was adjusted to 1.0 McFarland turbidity. Then the adjusted suspension with a turbidity of 1.0 McFarland was further serially diluted, resulting in tubes containing 108, 106, 104, and 102 CFU/mL equivalents [10]. Ten microlitre of suspension from each dilution was mixed with 2 mL sample reagent and incubated for 15 min at room temperature with intermittent shaking. Following the pretreatment, 2 mL of the sample was transferred to the cartridge. The results were automatically generated by GeneXpert assay after 90 min of the amplification reactions. The standard MTB H37Rv strain (ATCC 27294) and a clinical isolate with an amino acid substitution of Ser to Leu at codon 531 of rpoB (C017561) were used as positive quality controls, while the sterilized water for serial dilution was used to perform GeneXpert MTB/RIF experiments in parallel as negative control. All the tests were performed in triplicate.
Results
The GeneXpert results of different mycobacterial species were summarized in Table 1. Out of 12 NTM species, 5 were identified as MTB by GeneXpert at a bacterial load of 106, while the GeneXpert assay detected no MTB for the other 7 species. Notably, two species, including M. abscessus and M. smegmatis, were flagged as RIF-resistant MTB due to the high C(t) value of Probe E (Fig. 1). In addition, the detection failure of Probe D and Probe E was observed in M. phlei, resulting in indeterminate RIF resistance by GeneXpert. Both M. marinum and M. aurum had the amplification curves at all five probes, which were identified as RIF susceptible. When the further diluted samples were used, M. marinum was also detected as RIF-susceptible MTB by the GeneXpert testing at a bacterial load of 104. In addition, only MTB provided a positive result if the 102 of bacteria were loaded, indicating that the limit of detection of the GeneXpert assay was ~102 CFU/mL. We continued to analyze the amplification curves of different NTM species. As shown in Fig. 1, our results revealed that M. abscessus, M. smegmatis and M. phlei had no obvious exponential phase of the amplification plot, which might be associated with the mismatch between the probe and the chromosomal DNA target.
Discussion
GeneXpert has revolutionized the diagnostic algorithm for tuberculosis in the resource-limited settings. However, our findings have firstly demonstrated that the GeneXpert assay could misdiagnose some NTM species as MTB at a high bacterial load, which give important hints on the interpretation of GeneXpert results in the clinical practice. The identified “false-positive” results of several NTM species were associated with high C(t) values and absence of normal amplification plot. In addition to exhibit a strong correlation with RIF susceptibility, rpoB is a highly conserved housekeeping gene with little genetic diversity amongst various mycobacterial species [11]. Therefore, the highly conservative property of rpoB gene is associated with reducing the detection specificity of GeneXpert assay, especially for the samples with high bacterial load.
Notably, as a region with high TB burden, the proportion of NTM infections among all mycobacterial isolates in China has risen from 11.1% in 2000 to 22.9% in 2010, thus the epidemic of NTM in China has attracted more attention in recent years [9]. Out of the misdiagnosed NTM species in this study, M. abscessus is one of the major contributors causing NTM diseases in China. An epidemiology study from China reported that the infections of M. abscessus accounted for more than 20% of NTM pulmonary diseases [9]. Notably, due to the natural resistance to most anti-TB drugs, the clinical outcomes of patients with M. abscessus infection receiving the anti-TB regimens are not satisfactory [12]. There is no doubt that the misdiagnosis of M. abscessus as RIF-resistant MTB, an indicator of MDR-TB, will guide the clinicians to generate the improper therapeutic regimens, thereby resulting in the treatment failure. More importantly, the treatment of MDR-TB requires more toxic, less potent second-line drugs, as of today, takes up to 24 months of therapy according to current WHO recommendations [13]. In addition to unsatisfactory clinical outcomes, the adverse damage related to long-term administration of anti-MDR-TB treatment could be anticipated in these misdiagnosed patients. Because the mismatch between the probe and the target deeply relies on the bacterial load, the false-positive signals occur may be found in the clinical sputum with high smear positivity grade (3+ or 4+ grade according the national guideline for TB laboratory of China) [14]. Although the proportion of sputum with high positivity grade is unknown, the high prevalence of M. abscessus infection in China highlights the urgent need to develop strategies to prevent the emergence of misdiagnosis and treatment failure of M. abscessus diseases.
We try to find a potential solution for this intrinsic problem of GeneXpert assay. On one hand, Theron and colleagues reported that the use of GeneXpert assay exhibited excellent performance to predict the sputum smear status of TB patients [15]. On the other hand, in view of the inexact matching, the efficiency of the PCR reactions is significantly diminished, and the C(t) values of false-positive signals are also increased when compared with the actual bacterial load in the cartridge. Consistent with this hypothesis, we found that the C(t) values of NTM species with false-positive signals at a high bacterial load (equal to the sputum with a positivity grade of ~3+) were more than 30 cycles, while the C(t) values of MTB at the same bacterial load were about 15 cycles. Hence, the comparison of results between smear microscopy and GeneXpert, especially for the sputum specimens with a high positivity grade, provides a feasible method to monitor and solve the false-positive GeneXpert results due to NTM infections.
We also acknowledged several obvious limitations of this study. First, all the results of this study were obtained on the basis of artificial non-human samples, whereas the GeneXpert assay was optimized for human clinical samples. Further studies will be performed to evaluate the potential influence of misdiagnosis of MTB associated with NTM by GeneXpert in the clinical samples. Second, the update version of GeneXpert, known as the “Xpert MTB/RIF Ultra”, could yield improved performance in detecting MTB in clinical samples compared with conventional version. Unfortunately, this new version is not accessible in China. A better analysis would have been based on the new improved version of the GeneXpert assay in the future. Third, data based on this single study need to be replicated by colleagues in the field, as they would have a great impact on the user of GeneXpert.
In conclusion, our data have demonstrated that the presence of high C(t) value and abnormal amplification plot of GeneXpert results may be associated with high bacterial load among specific NTM cases. In view of widespread use of this assay, our data provide important hints on the interpretation of potential “false-positive” GeneXpert results. Further studies are urgently needed to confirm our results among the clinical sputum specimens from other countries.
References
Theron G, Peter J, Calligaro G, Meldau R, Hanrahan C, Khalfey H, et al. Determinants of PCR performance (Xpert MTB/RIF), including bacterial load and inhibition, for TB diagnosis using specimens from different body compartments. Sci Rep. 2014;4:5658.
Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, Krapp F, et al. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med. 2010;363:1005–15.
Moure R, Munoz L, Torres M, Santin M, Martin R, Alcaide F. Rapid detection of Mycobacterium tuberculosis complex and rifampin resistance in smear-negative clinical samples by use of an integrated real-time PCR method. J Clin Microbiol. 2011;49:1137–9.
Drobniewski F, Nikolayevskyy V, Balabanova Y, Bang D, Papaventsis D. Diagnosis of tuberculosis and drug resistance: what can new tools bring us? Int J Tuberc Lung Dis. 2012;16:860–70.
World Health Organization. Xpert MTB/RIF assay for the diagnosis of pulmonary and extrapulmonary TB in adults and children: policy update. Geneva: World Health Organization; 2013.
Pang Y, Lu J, Wang Y, Song Y, Wang S, Zhao Y. Study of the rifampin monoresistance mechanism in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2013;57:893–900.
Helb D, Jones M, Story E, Boehme C, Wallace E, Ho K, et al. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J Clin Microbiol. 2010;48:229–37.
Hillemann D, Rusch-Gerdes S, Boehme C, Richter E. Rapid molecular detection of extrapulmonary tuberculosis by the automated GeneXpert MTB/RIF system. J Clin Microbiol. 2011;49:1202–5.
Zhang Z, Pang Y, Wang Y, Cohen C, Zhao Y, Liu C. Differences in risk factors and drug susceptibility between Mycobacterium avium and Mycobacterium intracellulare lung diseases in China. Int J Antimicrob Agents. 2015;45:491–5.
Zhang Z, Wang Y, Pang Y, Liu C. Comparison of different drug susceptibility test methods to detect rifampin heteroresistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2014;58:5632–5.
Lee H, Bang HE, Bai GH, Cho SN. Novel polymorphic region of the rpoB gene containing Mycobacterium species-specific sequences and its use in identification of mycobacteria. J Clin Microbiol. 2003;41:2213–8.
Mougari F, Guglielmetti L, Raskine L, Sermet-Gaudelus I, Veziris N, Cambau E. Infections caused by Mycobacterium abscessus: epidemiology, diagnostic tools and treatment. Expert Rev Anti Infect Ther. 2016;14:1139–54.
Tiberi S, De Lorenzo S, Centis R, Viggiani P, D’Ambrosio L, Migliori GB. Bedaquiline in MDR/XDR-TB cases: first experience on compassionate use. Eur Respir J. 2014;43:289–92.
World Health Organization. Toman’s Tuberculosis: Case detection, treatment and monitoring-questions and answers. 2004; Geneva: World Health Organization.
Theron G, Pinto L, Peter J, Mishra HK, van Zyl-Smit R, Sharma SK, et al. The use of an automated quantitative polymerase chain reaction (Xpert MTB/RIF) to predict the sputum smear status of tuberculosis patients. Clin Infect Dis. 2012;54:384–8.
Acknowledgments
This work was supported by the National Key Research Program of China (2014ZX10003002) and the National Natural Science Foundation of China (81401739). We would like to thank members of the National Tuberculosis Reference Laboratory at the Chinese Center for Disease Control and Prevention for their technical assistance.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Funding
This work was supported by the Major State Basic Research Development Program of China (2014CB744403) and the National Natural Science Foundation of China (81401739).
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This study was approved by the Ethic Committee of Chinese Center for Disease Control and Prevention.
Rights and permissions
About this article
Cite this article
Pang, Y., Lu, J., Su, B. et al. Misdiagnosis of tuberculosis associated with some species of nontuberculous mycobacteria by GeneXpert MTB/RIF assay. Infection 45, 677–681 (2017). https://doi.org/10.1007/s15010-017-1044-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s15010-017-1044-x