Abstract
Purpose
To determine the incidence of patients co-colonised or co-infected with methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant Enterococcus faecium or extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae in four German tertiary care hospitals.
Methods
This study was conducted at four tertiary care hospitals (all with >1,000 beds) in different geographic regions in Germany (Berlin in the east, Luebeck in the north, Freiburg in the southwest and Nuernberg in the southeast). Routine surveillance data on MRSA, vancomycin-resistant enterococci (VRE) and ESBL-producing bacteria were analysed from 2007 to 2009. Co-colonisation or co-infection was defined as a patient having positive cultures for at least two of the following resistant pathogens: MRSA, VRE faecium or different species of ESBL-producing Enterobacteriaceae within one calendar year.
Results
A total of 896,822 patients were analysed, of which 10,066 patients harboured MRSA, VRE faecium and/or ESBL-producing Enterobacteriaceae, and 542 patients co-harboured at least two of those resistant pathogens. In 2009, 7.6% of the MRSA patients, 13.7% of the VRE faecium patients and even 16.1% of the ESBL-producing Enterobacteriaceae patients were co-colonised or co-infected. The incidence of patients with co-infection or co-colonisation increased steadily from 5 (2007) to 7 per 10,000 patients (2009).
Conclusions
Patients harbouring ESBL-producing Enterobacteriaceae or VRE faecium had a higher risk of being co-colonised or co-infected compared to what was to be extrapolated from their overall incidence. This might be linked to their gastrointestinal reservoir and impracticality to decolonise the gut of resistant VRE and ESBL-producing Enterobacteriaceae.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In 2002, vancomycin-resistant Staphylococcus aureus (VRSA) was isolated from two patients from southeast Michigan in the US. Both patients were colonised with methicillin-resistant S. aureus (MRSA) and vancomycin-resistant enterococci (VRE). Molecular methods confirmed that the vanA gene had been transferred from VRE to MRSA. There have been only 11 well-characterised examples of VRSA so far, but the risk of the transfer of resistance genes has inspired numerous case–control studies, mainly in the US and China. The aim was to determine the risk factors of co-colonisation or co-infection with MRSA and VRE [1–7]. Although the epidemiology of extended-spectrum β-lactamases (ESBLs) has changed and infections with ESBL-producing bacteria have increased dramatically, only a few studies have assessed the co-colonisation rates of VRE or MRSA and ESBL [4, 8].
Because data on secular trends and patients with multiple multidrug-resistant pathogens from Europe are scarce, we aimed to: (1) determine the incidence of patients with co-occurrence of MRSA, VRE faecium or ESBL-producing Enterobacteriaceae in four German hospitals and (2) to describe the change in the prevalence of patients with co-occurrence of those pathogens over the last 3 years (2007–2009).
Methods
This study was conducted at four tertiary care hospitals (all with >1,000 beds) in different geographic regions in Germany (Berlin in the east, Luebeck in the north, Freiburg in the southwest and Nuernberg in the southeast). The characteristics are displayed in Table 1.
All patients with proven MRSA, VRE faecium or ESBL-producing Enterobacteriaceae from 2007 to 2009 were eligible to be included in the study population. Every patient was counted only once during a calendar year. Patients were included irrespective of infection or colonisation. Likewise, all clinical and screening cultures were included. Copy strains were excluded. The genotyping of resistant strains was only performed if an outbreak was suspected.
Definition of co-colonisation or co-infection and statistical analysis
Co-colonisation or co-infection was defined as a patient having positive cultures for at least two of the following resistant pathogens: MRSA, VRE faecium or different species of ESBL-producing Enterobacteriaceae within one calendar year. The term “ESBL and ESBL” refers to co-infection or co-colonisation with different species of ESBL-producing bacteria. Differences over time were measured by the incidence density test and the significance level was set at P < 0.05.
Surveillance of MRSA, VRE and ESBL-producing Enterobacteriaceae
Admission screening for MRSA in high-risk patients has been recommended since 2004 in all four hospitals. High-risk patients are defined as patients who have chronic open wounds, are undergoing dialysis, have been transferred from hospitals or institutions with a known high prevalence of MRSA, and/or are older than 60 years of age and have been admitted to an intensive care unit. Electronic flagging of patients with identified MRSA, VRE faecium and/or ESBL-producing Enterobacteriaceae has been installed in all hospitals. All flagged patients were screened at hospital readmission. There was no general admission screening for ESBL or for VRE faecium. However, in the case of an outbreak (e.g. the VRE faecium outbreak in the paediatric department of one of the hospitals), screening was recommended. The screening policies in the four hospitals were not audited for compliance over time.
Microbiologic methods
Different microbiological identification systems are established in each of the four hospitals. For the purpose of the study, the identification methods were essentially as follows: Columbia Agar with sheep blood (COL SB+; Oxoid, Wesel, Germany) was used to isolate enterococci from different clinical samples. Enterococcus faecium was identified by using standard microbiological methods, including API 20 Strep or VITEK® 2 (bioMérieux, Marcy l’Etoile, France). Antimicrobial susceptibility to vancomycin was determined by the Etest® quantitative minimum inhibitory concentration (AB Biodisk, Solna, Sweden). Susceptibility interpretations followed the guidelines proposed by the Clinical and Laboratory Standards Institute (CLSI). Specific vancomycin-resistant genotypes (vanA, vanB or vanC) were determined by polymerase chain reaction analysis (GenoType Enterococcus, Hain Lifescience, Nehren, Germany) in all but one centre (Luebeck).
For the selective isolation and presumptive identification of MRSA, chromID MRSA™ Agar (bioMérieux, Marcy l’Etoile, France) was used. Presumptive S. aureus colonies from clinical samples on Columbia Agar with sheep blood (COL SB+; Oxoid, Wesel, Germany) were confirmed by positive catalase and by the detection of clumping factor, protein A and capsular polysaccharides (Pastorex™ Staph-Plus, Bio-Rad, Marnes-la-Coquette, France). Methicillin resistance was determined either by susceptibility testing by the disk diffusion method using the CLSI guidelines or by the detection of penicillin-binding protein (PBP2) (PBP2′ Latex Agglutination Test; Oxoid, Wesel, Germany) and confirmed by detection of the mecA gene (GenoType Staphylococcus, Hain Lifescience, Nehren, Germany).
Enterobacteriaceae strains selectively cultured on MacConkey medium were further characterised with biochemical tests (BBL Enterotube II, Becton–Dickinson Diagnostic Systems, Heidelberg/Germany, or Vitek® 2, bioMérieux, Marcy l’Etoile, France). ESBL-producers were determined by screening for ESBL with class III cephalosporins (cefotaxime, ceftazidime) and confirmed either by disk approximation or by Etest® ESBL strip cefotaxime and cefotaxime plus clavulanic acid, and ceftazidime and ceftazidime plus clavulanic acid (AB Biodisk, Solna, Sweden).
Infection control
The infection control departments in all four hospitals recommended contact precautions for patients colonised or infected with MRSA, VRE and ESBL (single room placement in most cases or cohorting, wearing of gloves and gowns, and screening of roommates, i.e. nasal screening for MRSA and rectal screening for VRE or ESBL). Known patients with MRSA, VRE or ESBL were screened on admission. There were no quantitative data available on compliance with infection control. Since recommendations are not always put into daily practice, it is important to note that each new case of ESBL, VRE or MRSA was followed by a local audit of the infection control staff.
Results
From 2007 to 2009, a total of 896,822 patients were admitted to the four hospitals and analysed. Of them, 10,066 patients harboured MRSA, VRE faecium and/or ESBL-producing Enterobacteriaceae, corresponding to 10,622 non-duplicate isolates. The number of patients with MRSA decreased from 2007 to 2009 (from 2,462 to 2,186 to 2,034), whereas VRE faecium increased (from 159 to 277 to 423), as did ESBL (from 818 to 1,040 to 1,223). The total number of admitted patients was as follows: 292,102 (2007), 298,146 (2008) and 306,574 (2009).
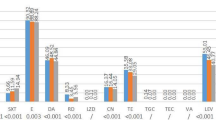
A total of 542 patients co-harboured MRSA, VRE faecium or ESBL-producing Enterobacteriaceae. In 2009, 7.6% of the MRSA patients (155 of 2,034) were co-colonised or co-infected and even 16.1% of the ESBL patients (230 of 1,223) and 13.7% of the VRE patients (58 out of 423). The number of co-infected or co-colonised patients increased from 157 to 219 over the last 3 years and accounted for 6% of all patients with MRSA, VRE faecium or ESBL-producing Enterobacteriaceae in 2009 (Fig. 1). The increase from 2007 to 2009 of all co-colonised or co-infected patients was statistically significant (P = 0.002). The majority of co-colonised or co-infected patients (62%) harboured MRSA and ESBL.
Number of patients co-colonised or co-infected with methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant Enterococcus faecium (VRE) or extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae in four German hospitals, 2007–2009. “ESBL and ESBL”: co-infection or co-colonisation with different species of ESBL-producing bacteria. The increase from 2007 to 2009 of all co-colonised or co-infected patients was statistically significant (P = 0.002).
The incidence of patients with co-infection or co-colonisation rose steadily by 40% from 5 (2007) to 7 per 10,000 patients (2009). The incidence of MRSA decreased from 84 to 66 per 10,000 patients. In contrast, VRE faecium burden almost tripled (from 5 to 9 to 14 VRE patients per 10,000 patients), indicating that 14 out of 10,000 patients were infected or colonised with VRE faecium in 2009. ESBL was 28 per 10,000 patients in 2007, increased to 35 in 2008 and was then 40 in 2009 (Fig. 2).
In one hospital, a VRE faecium outbreak in the paediatric department was suspected. Of 66 non-duplicate genotyped isolates, 53 belonged to one cluster, making an outbreak very likely (Table 2).
Generally, the distribution of pathogens in all patients with only one resistant pathogen (MRSA, VRE faecium or ESBL-producing Enterobacteriaceae) did not occur in parallel with the distribution in patients with co-infection or co-colonisation: patients with one resistant pathogen harboured MRSA in 66% of cases (2007–2009), but only in 37% of cases when only pathogens in co-infected or co-colonised patients were considered. In contrast, the distribution of VRE faecium or ESBL-producing Enterobacteriaceae in patients with more than one resistant pathogen was relatively higher than what was to be expected from the total distribution (Table 3).
Conclusions
The main findings are that: (1) 6 out of 10,000 admitted patients co-harboured methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant Enterococcus faecium (VRE) or extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae; (2) the incidence of co-infected or co-colonised patients increased from 5 to 7 per 10,000 patients over the last 3 years (2007–2009); and (3) 16.1% of the patients with ESBL-producing Enterobacteriaceae were co-colonised or co-infected in 2009. The co-occurrence of three different resistant pathogens (MRSA, VRE faecium or ESBL-producing Enterobacteriaceae) in patients is still rare.
Prior studies on co-colonisation or co-infection with MRSA and VRE demonstrated overall prevalence rates on admission ranging from 2.7 to 9.5% identified by clinical culture and active surveillance [2–4]. In one study in a US hospital, 28.6% of the VRE patients were co-colonised with MRSA as early as 1997 [7]. More recently, Reyes et al. [5] published co-carriage rates in patients with VRE and MRSA of 19.8%. In our study, the rate of co-carriage of MRSA and VRE faecium was about 13% (70 patients in 3 years).
We observed no vancomycin-resistant S. aureus (VRSA) in our cohort of almost 900,000 patients over the 3-year study period. Generally, the clinical importance of VRSA has been highlighted by the reports of VRSA occurring as a result of the transfer of the vanA gene from VRE [9]. However, it remains a mystery as to why the transfer of the vanA gene to S. aureus remains rare, although acquired vancomycin-resistant genes have been reported in MRSA and co-occurence is relevant.
With respect to Gram-negative bacteria, 4.7% co-carriage of MRSA and third-generation cephalosporin-resistant Gram-negative bacteria was reported in 1999 in ten Hong Kong intensive care units [4]. In two US intensive care units, 11% of the VRE patients were co-colonised with ESBL and 47% of the ESBL patients were co-colonised with VRE [8]. In this study, we found the predominant co-carriage to be MRSA and ESBL-producing Enterobacteriaceae (62% of patients with co-colonisation or co-infection). 16.1% of ESBL patients (230 of 1,223), 13.7% of VRE patients (58 out of 423) and only 7.6% of MRSA patients (155 of 2,034) were co-colonised or co-infected in 2009. We could confirm our initial hypothesis that the prevalence of patients with co-occurrence of resistant pathogens increases over time due to the general increase of patients with ESBL and VRE faecium. This is even true if all children with VRE faecium isolates from the department where an outbreak occurred are excluded. However, it is interesting to note that patients harbouring ESBL-producing Enterobacteriaceae or VRE faecium had a much higher risk to be co-colonised or co-infected than what was to be extrapolated from the general incidence. Further investigation is necessary in order to determine whether this higher rate of co-colonisation in comparison to MRSA is linked to the gastro-intestinal reservoir and impracticality of decolonising the gut of resistant enterococci and Enterobacteriaceae. The results of our study relied upon mostly clinical cultures. It can be expected that more cases of ESBL and VRE would be identified if a general admission screening were in place. The usefulness of general screening at admission, however, remains a matter of debate and its cost-effectiveness has yet to be proven [10]. According to Harris et al., isolating patients colonised by VRE would isolate 47% of the ESBL patients without the need for further testing [8]. The investigators hypothesised that testing for ESBL will be cost-effective only if patients were already colonised with VRE and only if infection control interventions would differ between those solely harbouring VRE and those who were co-colonised.
Our study has some limitations. First, our study took place under real-life conditions and, therefore, not all MRSA, VRE faecium or ESBL-producing Enterobacteriaceae were genotyped. This was done only when an outbreak was suspected. In one hospital, a VRE faecium outbreak took place in the paediatric department, inflating the incidence of patients harbouring VRE faecium. However, even if the patients from the outbreak are not considered in our calculation (one child in 2008 and four in 2009), the overall incidence of co-infected or co-colonised patients has still continuously increased. Furthermore, it cannot be ruled out that other outbreaks occurred but were not identified as such. In our view, the lack of complete typing data is a shortcoming, but our real-life data reflect, nevertheless, the situation which the hospitals have to deal with. Second, we cannot provide data on either the proportion of intensive care unit samples to medical and/or surgical samples nor a breakdown of the body site origin of the isolates. Third, the time period for the definition of co-colonisation or co-infection was a calendar year. Therefore, a patient colonised with MRSA in December and with VRE in January of the next year would not have fulfilled the definition. Fourth, the absence of general admission screening has to be taken into account. It cannot be ruled out that the four hospitals’ screening policies were heterogeneous because the screening policies in the four hospitals were not audited for compliance over time.
In conclusion, the burden of patients with co-colonisation or co-infection with MRSA, VRE faecium or ESBL-producing Enterobacteriaceae increased from 2007 to 2009 by 40%. This was especially the case in patients carrying ESBL-producing Enterobacteriaceae. Clinicians can expect to face increasing numbers of patients with co-occurrence of resistant pathogens. Therefore, three points are paramount: prevention of transmission, limiting antibiotic use to curtail the selection and persistence of predominant clones, and developing strategies to influence carriage, especially in the intestinal tract as an important reservoir of antibiotic resistance genes.
References
Han SH, Chin BS, Lee HS, Jeong SJ, Choi HK, Kim CK, Kim CO, Yong D, Choi JY, Song YG, Lee K, Kim JM. Recovery of both vancomycin-resistant enterococci and methicillin-resistant Staphylococcus aureus from culture of a single clinical specimen from colonized or infected patients. Infect Control Hosp Epidemiol. 2009;30:130–8.
Warren DK, Nitin A, Hill C, Fraser VJ, Kollef MH. Occurrence of co-colonization or co-infection with vancomycin-resistant enterococci and methicillin-resistant Staphylococcus aureus in a medical intensive care unit. Infect Control Hosp Epidemiol. 2004;25:99–104.
Furuno JP, Perencevich EN, Johnson JA, Wright MO, McGregor JC, Morris JG Jr, Strauss SM, Roghman MC, Nemoy LL, Standiford HC, Hebden JN, Harris AD. Methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci co-colonization. Emerg Infect Dis. 2005;11:1539–44.
Ho PL. Carriage of methicillin-resistant Staphylococcus aureus, ceftazidime-resistant gram-negative bacilli, and vancomycin-resistant enterococci before and after intensive care unit admission. Crit Care Med. 2003;31:1175–82.
Reyes K, Malik R, Moore C, Donabedian S, Perri M, Johnson L, Zervos M. Evaluation of risk factors for coinfection or cocolonization with vancomycin-resistant enterococcus and methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2010;48:628–30.
Wang Z, Cao B, Liu YM, Gu L, Wang C. Investigation of the prevalence of patients co-colonized or infected with methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci in China: a hospital-based study. Chin Med J. 2009;122:1283–8.
Franchi D, Climo MW, Wong AH, Edmond MB, Wenzel RP. Seeking vancomycin resistant Staphylococcus aureus among patients with vancomycin-resistant enterococci. Clin Infect Dis. 1999;29:1566–8.
Harris AD, Nemoy L, Johnson JA, Martin-Carnahan A, Smith DL, Standiford H, Perencevich EN. Co-carriage rates of vancomycin-resistant Enterococcus and extended-spectrum beta-lactamase-producing bacteria among a cohort of intensive care unit patients: implications for an active surveillance program. Infect Control Hosp Epidemiol. 2004;25:105–8.
Chang S, Sievert DM, Hageman JC, Boulton ML, Tenover FC, Downes FP, Shah S, Rudrik JT, Pupp GR, Brown WJ, Cardo D, Fridkin SK. Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N Engl J Med. 2003;348:1342–7.
Meyer E, Serr A, Schneider C, Utzolino S, Kern WV, Scholz R, Dettenkofer M. Should we screen patients for extended-spectrum beta-lactamase-producing enterobacteriaceae in intensive care units? Infect Control Hosp Epidemiol. 2009;30:103–5.
Acknowledgements
We thank Ryan Plocher for his help in preparing the manuscript.
Conflict of interest
All authors: none to declare.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Meyer, E., Ziegler, R., Mattner, F. et al. Increase of patients co-colonised or co-infected with methicillin-resistant Staphylococcus aureus, vancomycin-resistant Enterococcus faecium or extended-spectrum β-lactamase-producing Enterobacteriaceae . Infection 39, 501–506 (2011). https://doi.org/10.1007/s15010-011-0154-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s15010-011-0154-0