Abstract
Mercury (Hg) is one of the major toxic heavy metals because it bioaccumulates and biomagnifies in animal and human bodies via the food chain. To eliminate heavy metal contamination, plants are being used as removal agents of pollutants/toxic chemicals from the environment. The present study was mainly focused on elucidating the potential phytotoxic effects of Hg heavy metal ion exposure on Sesbania grandiflora seedlings. Growth of seedlings was significantly affected (56 %) at 60 mg L−1 Hg concentration. The level of chlorophyll pigment contents was increased in Hg-treated plants compared to the control. Malondialdehyde content and antioxidative enzyme activities were found to be significantly increased by increasing the concentration of Hg exposure up to 40 mg L−1 while slightly decreased at higher doses. The DNA alterations appearing in the random amplified polymorphic DNA (RAPD) profiles of leaf and root tissues following Hg heavy metal exposure included the disappearance of normal DNA bands and the appearance of new bands compared to the untreated controls. This result strongly indicated that genomic template stability was significantly affected by Hg-induced stress in S. grandiflora seedlings. It is concluded that DNA polymorphisms detected by RAPD fingerprinting analysis could be used as potential molecular markers for the evaluation of Hg heavy metal ion-induced genotoxic effects in other plant species.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Soil and water contamination with heavy metals has become a worldwide problem leading to losses in agricultural crops yield and causes hazardous health effects to human population (Taghinia Hejabi et al. 2010; Conceição et al. 2013; Moaref et al. 2014). Mercury is a strong phytotoxic heavy metal ion that causes inhibition of plant growth as well as long-term effects on soil fertility. Mercury (Hg) is one of the major toxic metals which enter into the food chain (Suszcynsky and Shan 1995). The main sources of mercury pollution include mining of gold and silver, the coal industry, untreated discarded batteries, and industrial waste disposal (Pilon-Smits and Pilon 2000).
High concentration of mercury ions induces oxidative stress by increasing the formation of reactive oxygen species (ROS) like superoxide radical (O· −2 ), singlet oxygen (1O2), hydroxyl radical (·OH), and hydrogen peroxide (H2O2) in plant cells, which are responsible for peroxidase damages to fatty acids, nucleic acids, protein, and chlorophyll contents (Gallego et al. 2002). However, plant cells have a complex antioxidant system capable of scavenging ROS by redox homeostasis. The antioxidant system comprises, among other components, enzymes such as superoxide dismutase (SOD), catalase (CAT), ascorbate peroxidase (APX), and peroxidase (POX) (Noctor and Foyer 1998). SOD is a major ROS scavenger, and its enzymatic action results in H2O2 and O2 formation, while CAT and peroxidase enzymes are involved in scavenging H2O2. Under heavy metal stress conditions, it has been shown that the level of detoxifying enzyme activities was increased in plants due to the enhanced production of ROS, which affects cell membrane structure, nucleic acids, and photosynthetic pigment contents (Zhang et al. 2007). Malondialdehyde (MDA) is considered a cytotoxic product of lipid peroxidation and an indicator of free radical production in stressed cells (Hu et al. 2012; Li et al. 2013). Therefore, enhanced activities of antioxidative enzymes in hyperaccumulator plants may be considered as typical defense components against heavy metal stress-induced toxicity.
Heavy metal-induced genotoxicity on genomic DNA molecules may be detected by applying traditional polymerase chain (PCR) assays to identify mutations and DNA damage (Cenkci et al. 2009; Liu et al. 2009). However, mutations may inhibit the annealing of the oligonucleotide primers, while DNA damage may affect the Taq DNA polymerase activity, which ultimately changes the number of newly amplified DNA bands (Atienzar and Jha 2004). This problem can be avoided by applying the RAPD fingerprinting method, which is based on the PCR amplification of total genomic DNA with multiple annealing of 10mer random arbitrary primers (Quintela-Sabaris et al. 2005; Hagger et al. 2006). The PCR amplifications thus produce multiple DNA bands with variable sizes (50–450 bp) which can be detected by agarose gel electrophoresis to establish RAPD fingerprints (Liu et al. 2009). The RAPD technique has been successfully applied to detect genomic sequence changes induced by several DNA damaging agents, such as Pb (Cenkci et al. 2009), Hg (Cenkci et al. 2010), Cd (Liu et al. 2009), As (Ahmad et al. 2012), and 2,4-dichlorophenoxyacetic acid (Aksakal et al. 2013) in plants. Although there are a few reports on heavy metal stress-induced genotoxic effects in plants, mercury ion-induced genotoxicity requires additional research to identify the exact mechanism under stressed conditions.
Hyperaccumulator plants represent a resource for phytoremediation of heavy metal-polluted sites, as they can tolerate, uptake, and translocate heavy metals into their biomass at levels toxic to most organisms (Zavoda et al. 2001). Mercury accumulation has been studied in various plant species including Medicago sativa (Zhou et al. 2007), Pteris vittata and Nephrolepis exaltata (Chen et al. 2009), Sesbania drummondii (Venkatachalam et al. 2009), Jatropha curcas (Gao et al. 2010), Oriza sativa (Meng et al. 2010), and Mentha arvensis (Manikandan and Venkatachalam 2011). Sesbania grandiflora a growing tree species with high biomass about which the effect of Hg exposure on physiochemical and molecular changes was previously unknown. This research work was carried out from February 2013 to November 2013 in the Plant Genetic Engineering and Molecular Biotechnology Lab, Department of Biotechnology, Periyar University, TN, India. In the present study, we hypothesized that (1) if enhanced levels of photosynthetic pigments, MDA content, and antioxidative enzymes level in S. grandiflora seedlings differ in sensitivity to Hg exposure, (2) then to induce genotoxicity by causing genomic DNA changes, an efficient detoxification mechanism is evolved to overcome the heavy metal-induced oxidative stress. The results of the present study also demonstrate the effect of Hg exposure in S. grandiflora: the species capability to activate a multi-defense strategy against oxidative damage and genotoxicity caused by Hg ions, which may be a key factor in the detoxification mechanism of tolerance to unfavorable conditions.
Materials and methods
Plant material and growth conditions
Sesbania grandiflora seeds were germinated on trays containing a mixture of coir waste and sand in a 3:1 ratio. After germination, seedlings (10-day-old) were grown under a hydroponic system containing 500 ml of Hoagland nutrient solution (Hoagland and Arnon 1950) and were continuously aerated. After 7 days in the hydroponics culture, seedlings were treated with different concentrations of HgCl2 (10, 20, 30, 40, 50, and 60 mg L−1) for 10 days. Seedlings without mercury treatment served as controls with triplicate maintained for each treatment. After 10 days of treatment, root and shoot samples were harvested separately and measured for various physiochemical and molecular changes.
Determination of growth inhibitory rate, biomass, and relative water content
The length of the longest root and shoot of each seedling was recorded after the 10-day treatment period. Root and shoot lengths of S. grandiflora seedlings were measured, and growth inhibitory rate (GIR %) of roots and shoots was calculated by using the formula (1 − x/y) × 100, where x was the average value detected in the control and y was one in each sample treated (Korpe and Aras 2011).
For biomass and relative water content (RWC) analysis, plants were again separated into roots and shoots. Wet plant biomass (FW) was immediately determined. The samples were dried in a hot air oven for 48 h at 65 °C for determination of dry weight (DW). The RWC was calculated as, RWC (%) = [(FW − DW)/FW] × 100 (Chen et al. 2009).
Quantification of mercury contents and translocation factor
Plant tissues collected from the 60 mg L−1 mercury treatment were dried in a hot air oven at 65 °C for 48 h to remove all the moisture content. The oven-dried tissues were ground into a fine powder and used for metal analysis by ICP-MS. The level of mercury content in root and shoot tissues was quantified according to the method of Israr and Sahi (2006). Translocation factor (TF) was calculated as described by Marchiol et al. (2004) as
Estimation of chlorophyll pigment contents
Chlorophyll pigments were extracted from fresh leaf samples using 80 % (v/v) acetone and chlorophyll a, b, and carotenoid contents were estimated spectrophotometrically at 665, 649, and 470 nm according to the method of Lichtenthaler (1987) and expressed in mg−1 g FW.
Measurement of MDA contents
Lipid peroxidation was determined by measuring the total amount of malondialdehyde (MDA) contents as described by Davenport et al. (2003). Briefly, fresh root and leaf tissues (0.2 g) were homogenized using 2 ml of 5 % (w/v) trichloroacetic acid in an ice bath and centrifuged at 10,000 rpm for 10 min at 4 °C. About 2 ml supernatant was mixed with 2 ml of 0.67 % (w/v) thiobarbituric acid, incubated in a boiling water bath for 30 min, then cooled and centrifuged. The absorption of supernatant was carried out at 450, 532, and 600 nm. The MDA content was calculated as
where Vt = 0.0021; W = 0.2 g.
Determination of antioxidative enzyme activities
Both root and leaf tissues (>200 mg) from S. grandiflora seedlings were homogenized separately in a pre-chilled mortar and pestle under ice-cold conditions with 2.0 ml of extraction buffer [50 mM phosphate buffer (pH 7.5), 0.5 mM ascorbate, and 1 mM EDTA]. The homogenate was centrifuged at 10,000 rpm for 15 min. The supernatant was used for the measurement of SOD (Beauchamp and Fridovich 1971), CAT (Beer and Sizer 1952), APX (Nakano and Asada 1987), and POX (Zhang et al. 1995) antioxidative enzyme activities. In addition, the protein content was also measured according to the method of Bradford (1976) using bovine serum albumin (BSA) as a standard.
The SOD activity was estimated by measuring its ability to inhibit the photochemical reduction of nitroblue tetrazolium (NBT) (Beauchamp and Fridovich 1971). The catalase (CAT) activity was determined by measuring the decomposition of hydrogen peroxide (Beer and Sizer 1952). The APX activity was determined according to the method of Nakano and Asada (1987). The POX activity was estimated as per the method described by Zhang et al. (1995). Total protein content was estimated according to the method of Bradford (1976) using bovine serum albumin (BSA) as standard.
Genomic DNA isolation and RAPD–PCR analysis
Total cellular genomic DNA was isolated from root and leaf tissues of S. grandiflora seedlings by the CTAB extraction method (Doyle and Doyle 1990). Fresh leaf and root tissues (100 mg) were obtained from 10, 20, 40, and 60 mg L−1 mercury-treated seedlings along with control (untreated) seedlings. Briefly, samples were ground into fine powder and mixed with 2× CTAB buffer. The extract was mixed with equal volume of phenol–chloroform–isoamyl alcohol (25:24:1) and centrifuged at 8000 rpm for 10 min. The supernatant was re-extracted with equal volume of chloroform and isopropyl alcohol (24:1). After centrifugation, DNA was precipitated by adding 0.6 ml of isopropanol and the DNA pellet was carefully washed with 70 % (v/v) ethanol and air-dried. It was dissolved in TE buffer and used for PCR amplification. RAPD–PCR amplifications were carried out according to the method of Williams et al. (1990). DNA amplifications were performed in 20 μl reaction volume, which contained 100 mM Tris–HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 1.5 mM dNTP, 0.5U Taq DNA polymerase enzyme, 25 ng of template DNA, and 25 nM of RAPD primer (Operon Technologies Inc, CA, USA). The PCR amplification profile consisted of a first cycle at 94 °C for 4 min, followed by 40 cycles at 94 °C for 1 min, 37 °C for 1.5 min, 72 °C for 2 min, and final extension at 72 °C for 7 min. Amplicons were analyzed by agarose gel electrophoresis, and the reproducibility of the amplified products was tested twice for each experiment.
Estimation of genomic template stability
Disappearance of normal bands and appearance of new bands in RAPD profiles (i.e., polymorphisms), as compared to the control profile, were scored as ‘0’ and ‘1,’ respectively.
Statistical analysis
Data in table and figures are mean values of three independent replicates. Significant differences among treatments were analyzed by one-way ANOVA, taking p ≤ 0.05 at significance level, using Graphpad InStat Software.
Results and discussion
Effect of mercury treatment on plant growth, biomass, and relative water content
The major responses of plants to Hg heavy metal-induced toxicity included reduction in seedling growth, enhancement of antioxidative stress enzyme activities, and alteration of genomic DNA. S. grandiflora seedlings exposed to different concentrations of HgCl2 exhibited inhibition of both root and shoot growth, with shoots affected more than roots after 10 days of treatment. The reduction in seedling growth was 47 and 56 % for shoots and roots, respectively (Table 1). Hg-treated plants did not show any visible toxicity symptoms up to 40 mg L−1 Hg. However, toxicity symptoms like chlorosis, drying of leaf edges, browning of roots, and dropped leaves were seen in higher Hg concentrations compared to the controls. Plant growth was significantly affected by Hg heavy metal toxicity. Plant tolerance to heavy metal-induced stress was estimated by the percent of root and/or shoot growth inhibition by heavy metal ions (Wang and Zhou 2005). Similar observations were also recorded under Hg stress in Sesbania drummondii (Venkatachalam et al. 2009) and Mentha arvensis (Manikandan and Venkatachalam 2011) seedlings.
The growth of S. grandiflora seedlings was assessed in terms of biomass accumulation, which was significantly affected at higher concentrations of Hg. Biomass was reduced 19 % in seedlings grown at 60 mg L−1 Hg compared to the controls. Growth rate was also decreased with Hg exposure, as expected, due to similar findings in studies by Venkatachalam et al. (2009) and Gao et al. (2010). Accumulation of Hg heavy metal ions in plant cells may trigger water deficit, resulting in reduced growth and development of plants (Gao et al. 2010).
Relative water content (RWC), therefore, is an indicator for phytotoxicity The RWC of our samples was decreased by 5 % at 60 mg L−1 Hg treatment compared to control seedlings (Table 1). These results are comparable to those reported for Pisum sativum (Malecka et al. 2009) and Jatropha curcus seedlings (Gao et al. 2010).
Mercury accumulation in S. grandiflora seedlings
An efficient Hg accumulation mechanism in roots could represent a new and interesting phenomenon for the development of phytoremediation strategies in which a higher concentration of the pollutants remains tightly adhered to the plant tissues. Hg accumulation has also been found to be higher in roots than in shoots of Jatropha curcas (Gao et al. 2010) and Atriplex codonocarpa (Lomonte et al. 2010). The potential for Hg ions hyperaccumulation by S. grandiflora seedlings clearly indicates that this plant species may have an efficient physiological mechanism for the removal of heavy metal ions from contaminated sites or water bodies.
Effect of mercury stress on chlorophyll pigment contents
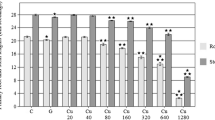
The total chlorophyll and carotenoid contents in leaves of S. grandiflora seedlings increased with increasing Hg dose compared to the control seedlings, though slightly decreased at higher concentrations. Seedlings treated with 60 mg L−1 Hg showed 92 % higher chlorophyll a, 76 % higher chlorophyll b, and 69 % higher carotenoid contents over the controls (Fig. 1). The photosynthetic pigment content changes may be attributed to an increased level of chlorophyll synthesis by mercury stress, as a similar result was reported in chromium (Cr)-treated Pisitia stratiotes (Sinha et al. 2005) and iron (Fe)-treated Bacopa monnieric (Sinha and Saxena 2006). It is believed that heavy metal-hyperaccumulating plants produce a higher level of photosynthetic pigment contents, which may be a potential detoxification mechanism for the efficient removal of metal ions, including Hg, from contaminated sites.
Effect of Hg treatment on lipid peroxidation
The antioxidant enzymes and certain metabolites important in alleviating heavy metal-induced oxidative damage in plants were monitored. Lipid peroxidation in leaves and roots of S. grandiflora seedlings, measured as MDA content, increased with increasing Hg concentration up to 40 mg L−1 and was slightly decreased at higher doses (Fig. 2). The total MDA content observed was 60.25 and 58.45 % for leaf and root tissues, respectively, as compared to controls. MDA is the decomposed product of polyunsaturated fatty acids of biomembranes, and its increase when plants are under Hg stress is an indicator of the extent of oxidative stress (Hou et al. 2007; Hu et al. 2012). Mercury treatment induces a higher level of ROS production in chloroplasts and peroxisomes of plants. These ROS can cause severe lipid peroxidation due to the removal of hydrogen ions from unsaturated fatty acids leading to the generation of lipid radicals and reactive aldehydes (Chen et al. 2009). This suggests that the biomembrane was damaged due to Hg-induced oxidative stress in S. grandiflora seedlings. An increased level of lipid peroxidation may be attributed to the enhanced activity of antioxidative enzymes to reduce H2O2 levels, in Hg-treated plants; therefore, the cell membrane damage could be minimized (Zhang et al. 2007).
Effect of Hg-induced stress on antioxidative enzyme activity
Hg-induced stress triggered some of the key antioxidant defense enzyme systems involved in H2O2 detoxification mechanisms in S. grandiflora seedlings. Superoxide dismutase is considered a first defense system against ROS as it acts on superoxide free radicals, which are generated in different compartments of the cell (Alscher and Erturk 2002).
The SOD activity was positively correlated with Hg treatment compared to the control, while it was slightly decreased at higher dose. Maximum level of SOD activity increased by 196 and 625 % for root and leaf tissues, respectively (Fig. 3a). Feng-tao et al. (2013) described that cadmium heavy metal treatment enhanced SOD activity in Hibiscus cannabinus plant. The decreased level of SOD activity, at higher concentration Hg treatment, might be attributed to enzyme damage from the excess production of free radicals and peroxisomes, likely due to the deleterious effect of excess H2O2 or its poisonous ROS derivatives. CAT eliminates H2O2 by breaking it down directly to form water and oxygen. A concentration-dependent increase in catalase activity was detected up to 40 mg L−1 Hg, which then slightly decreased at higher doses (Fig. 3b). However, CAT activity increased 16 and 66 % for root and leaf tissues, respectively, over the controls. Contrary to our results with S. grandiflora, decline in the activity of catalase with increased heavy metal concentrations was reported for Becopa monneri (Mishra et al. 2006) and Lemna gibba (Parlak and Yilmaz 2013). It is likely that over production of ROS by heavy metal-induced stress can inactivate CAT activity at higher doses of heavy metal ions, probably by inactivating the enzyme-bound heme group (Willekens et al. 1997).
APX activity was significantly enhanced with increasing Hg concentration up to 40 mg L−1 and slightly decreased at higher doses. APX activity in Hg-treated seedlings increased 28 and 34 % for root and leaf tissues, respectively, at 40 mg L−1 Hg over the control seedlings (Fig. 3c). An increased level of APX activity may effectively scavenge H2O2 to protect plant cells against oxidative damage under mercury heavy metal ion-induced stress. Similar results were also reported by Feng-tao et al. (2013).
POX activity was positively correlated with Hg treatment, though slightly decreased at higher concentrations (Fig. 3d). We detected a 67 and 35 % increase in POX activity for leaf and root tissues, respectively, over control seedlings. POX could act as an effective quencher of reactive oxygen species and peroxy radicals produced by heavy metal exposure in plants (Radotic et al. 2000). POX consumes H2O2 to generate phenoxy compounds that are polymerized to produce cell wall components such as lignans (Hu et al. 2012). The positive effect of heavy metal exposure on the antioxidative defense system has also been demonstrated in numerous other studies (Zhang et al. 2007; Zhou et al. 2007; Gao et al. 2010; Feng-tao et al. 2013). The activity of the antioxidative enzymes SOD, CAT, APX, and POX were all found to be higher in roots than in leaf tissues under mercury heavy metal-induced stress, clearly indicating that both the root and leaf tissues of S. grandiflora have evolved efficient detoxification mechanisms to survive under mercury toxicity in the hydroponic system.
Effect of Hg on RAPD profiles
RAPD fingerprinting analysis of DNA banding patterns showed clear differences between untreated control and Hg-treated S. grandiflora seedlings, with distinct changes (absence and/or presence) in the number and size of PCR amplicons with each primer. A total of 60 random oligonucleotide primers were screened for PCR amplification in which about 11 primers produced RAPD banding pattern and only five primers amplified clear, scorable, and reproducible DNA fragments (Fig. 4). In the RAPD banding pattern, few DNA fragments were absent while additional DNA bands were amplified in the Hg-exposed samples. Intensity of the newly amplified DNA bands varied with the concentration of Hg treatments, with a decreased DNA band intensity observed at higher concentrations.
RAPD profiles generated using DNA samples isolated from the leaf and root tissues of control S. grandiflora seedlings and those treated with different concentrations (10, 20, 40, and 60 mg L−1) of mercury (Hg) metal ion. ↘ appearance of new bands, ↖ disappearance of normal bands, M marker (DNA ladder), CL control leaf, CR control root
Changes in band intensity and loss of bands may be due to the modification of oligonucleotide priming sites mainly caused by genomic rearrangements, or potentially the occurrence of point mutations or DNA damage in the binding sites (Cenkci et al. 2010). Amplification of new bands could be attributed to the presence of priming sites, which become accessible to oligonucleotide primers after structural changes in DNA sequences have occurred due to mutations (resulting in new annealing events), large deletions (bringing two preexisting annealing sites closer), or homologues recombinations (Liu et al. 2009; Korpe and Aras 2011). Earlier studies described that the appearance of new DNA bands and disappearance of normal DNA bands could be due to the mutation and/or DNA damage caused by heavy metals (Liu et al. 2009; Cenkci et al. 2010). Genomic template stability in S. grandiflora seedlings was significantly affected by Hg heavy metal toxicity-induced oxidative stress. Alterations observed in RAPD banding pattern could be modifications in genomic template stability (GTS), which could be directly compared with changes in various parameters such as physiological growth rate, chlorophyll pigment content, and antioxidative enzyme activity (Cenkci et al. 2010; Korpe and Aras 2011).
Conclusion
Accumulation of Hg heavy metal ions in plant tissues induced both physiochemical and molecular changes in S. grandiflora seedlings. Hg heavy metal ions induced severe oxidative damage as evidenced by enhanced level of lipid peroxidation, indicating the existence of toxic ROS. Thus, S. grandiflora has the detoxification potential to scavenge excess ROS efficiently via coordinated activation of SOD, CAT, APX, and POX antioxidative defense systems. Furthermore, the DNA changes detected by our RAPD assay could be used as a molecular tool to identify the genotoxic effect of heavy metal toxicity-induced stress in plants. An efficient hyperaccumulation mechanism of Hg metal ions in S. grandiflora roots could represent a novel and interesting mechanism for the establishment of phytoremediation models that require a large quantity of the pollutants remain tightly bound to plant tissues. Our results strongly indicate that Hg ion was retained by root cells of S. grandiflora, confirming that this species has a heavy metal hyperaccumulation mechanism, and shows potential as a phytoextracting species via the rhizofiltration method in the polluted environment.
References
Ahmad MA, Guar R, Gupta M (2012) Comparative biochemical and RAPD analysis in two varieties of rice (Oryza sativa) under arsenic stress by using various biomarkers. J Hazard Mater 217–218:141–148
Aksakal O, Erturk FA, Sunar S, Bozari S, Agar G (2013) Assessment of genotoxic effects of 2,4-dichlorophenoxyacetic acid on maize by using RAPD analysis. Ind Crops Prod 42:552–557
Alscher RG, Erturk NL (2002) Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. Environ Exp Bot 53:1331–1341
Atienzar FA, Jha AN (2004) The random amplified polymorphic DNA (RAPD) assay to determine DNA alterations, repair and transgenerational effects in B(a)P exposed Daphnia magna. Mutat Res 552:125–140
Beauchamp C, Fridovich I (1971) Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal Biochem 44:276–287
Beer JRF, Sizer IW (1952) A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J Biol Chem 195:133–140
Bradford MM (1976) A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein binding. Anal Biochem 72:248–254
Cenkci S, Yildiz M, Cigerci IH, Konuk M, Bozdag A (2009) Toxic chemicals-induced genotoxicity detected by random amplified polymorphic DNA (RAPD) in bean (Phaseolus vulgaris L.) seedlings. Chemosphere 76:900–906
Cenkci S, Cigerci IH, Yildiz M, Ozay C, Bozdag A (2010) Lead contamination reduces chlorophyll and genomic template stability in Brassica napa L. Environ Exp Bot 67:467–473
Chen J, Shiyab S, Han FX, Monts DL, Waggoner AW, Su ZY (2009) Bioaccumulation and physiological effects of mercury in Pteris vittata and Nephrolepis exaltata. Ecotoxicology 18:110–121
Conceição FT, Navarro GRB, Silva AM (2013) Anthropogenic influences on Cd, Cr, Cu, Ni, Pb and Zn concentrations in soils and sediments in a watershed with sugar cane crops at São Paulo State, Brazil. Int J Environ Res 7(3):551–560
Davenport SB, Gallego SM, Benavides MP, Tomarow ML (2003) Behaviour of antioxidant defense system in the adaptive response to salt stress in Helianthus annus L. J Plant Growth Regul 40:81–88
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Feng-tao LI, Jian-min QI, Gao-yang Z, Li-hui L, Ping-ping F, Fen TA, Jian-tang XU (2013) Effect of cadmium stress on the growth, antioxidative enzymes and lipid peroxidation in two kenaf (Hibiscus cannabinus L.) plant seedlings. J Int Agri 12:610–620
Gallego S, Benavides M, Tomarow A (2002) Involvement of an antioxidant defense system in the adaptive response to heavy metal ions in Helianthus annus L. cells. Plant Growth Regul 36:267–273
Gao S, Yang C, Tang L, Zhu J, Xu Y, Wang S, Chen F (2010) Growth and antioxidant responses in Jatropha curcas seedling exposed to mercury toxicity. J Hazard Mater 182:591–597
Hagger JA, Atienzar FA, Jha AN (2006) Genotoxic, cytotoxic, developmental and survival effects of tritiated water in the early life stages of the marine mollusc, Mytilus edulis. Aquatic Toxicol 74:205–217
Hoagland R, Arnon DI (1950) The water-culture method of growing plants without soil. Calif Agric Exp Station Circ 347:1–32
Hou W, Chen X, Song G, Wang Q, Chang CC (2007) Effects of copper and cadmium on heavy metal polluted water body restoration by duckweed (Lemna minor). Plant Physiol Biochem 45:62–69
Hu R, Sunc K, Suc X, Pana Y, Zhang Y (2012) Physiological responses and tolerance mechanisms to Pb in two xerophils: Salsola passerina Bunge and Chenopodium album L. J Hazard Mater 205–206:131–138
Israr M, Sahi SV (2006) Antioxidative responses to mercury in the cell cultures of Sesbania drummondii. Plant Physiol Biochem 44:590–595
Korpe DA, Aras S (2011) Evaluation of copper-induced stress on eggplant (Solanum melongena L.) seedlings at the molecular and population levels by use of various biomarkers. Mutat Res 719:29–34
Li B, Shi JB, Wang X, Meng M, Huang L, Qi XL, He B, Ye ZH (2013) Variations and constancy of mercury and methylmercury accumulation in rice grown at contaminated paddy field sites in three Provinces of China. Environ Pollut 181:91–97
Lichtenthaler HK (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148:350–382
Liu W, Yang YS, Li PJ, Zhou QX, Xie LJ, Han YP (2009) Risk assessment of cadmium-contaminated soil on plant DNA damage using RAPD and physiological indices. J Hazard Mater 161:878–883
Lomonte C, Sgherri C, Baker JM, Kolev SD, Navari-Izzo F (2010) Antioxidative responses of Atriplex codonocarpa to mercury. Environ Exp Bot 69:9–16
Malecka A, Piochalak A, Tomaszewska B (2009) Reactive oxygen species production and antioxidative defense system in pea root tissues treated with lead ions: the whole roots level. Acta Physiol Plant 31:1053–1063
Manikandan R, Venkatachalam P (2011) Risk assessment of mercury ion heavy metal exposure on physiological and biochemical changes and DNA damage using RAPD analysis in Mentha arvensis seedlings. Plant Cell Biotech Mol Biol 12:41–50
Marchiol L, Assolari S, Sacro P, Zerbia A (2004) Phytoremediation of heavy metals by Canola (Brassica napus) and radish (Raplanus sativus) grown on multi contaminated soil. Environ Pollut 132:21–27
Meng B, Feng X, Qiu G, Cai Y, Wang D (2010) Distribution patterns of inorganic mercury and methylmercury in tissues of rice (Oryza sativa L.) plants and possible bioaccumulation pathways. J Agric Food Chem 58:4951–4958
Mishra S, Srivastava S, Tripathi RD, Govindarajan R, Kuriakose SV, Prasad MNV (2006) Phytochelation synthesis and responses of antioxidants during cadmium stress in Bacopa monnieri L. Plant Physiol Biochem 44:25–37
Moaref S, Sekhavatjou MS, Hosseini Alhashemi A (2014) Determination of trace elements concentration in wet and dry atmospheric deposition and surface soil in the largest industrial city, Southwest of Iran. Int J Environ Res 8(2):335–346
Nakano Y, Asada K (1987) Purification of ascorbate peroxidase in spinach chloroplasts: its inactivation in ascorbate depleted medium and reactivation by monodehydroascorbate radical. Plant Cell Physiol 28:131–140
Noctor G, Foyer CH (1998) Ascorbate and glutathione: keeping active oxygen under control. Plant Physiol Plant Mol Biol 49:249–279
Parlak KC, Yilmaz DD (2013) Ecophysiological tolerance of Lemna gibba L. exposed to cadmium. Ecotoxicol Environ Saf 91:79–85
Pilon-Smits E, Pilon M (2000) Breeding mercury-breathing plants for environmental cleanup. Trends Plant Sci 5:235–236
Quintela-Sabaris C, Kidd PS, Fraga MI (2005) Identification of metalliferous ecotypes of Cistus ladanifer L. using RAPD markers. Z Naturforsch 60:229–235
Radotic K, Ducic T, Mutavdzic D (2000) Changes in peroxidase activity and isoenzymes in spruce needles after exposure to different concentrations of cadmium. Environ Exp Bot 44:105–113
Sinha S, Saxena R (2006) Effect of iron on lipid peroxidation, and enzymatic and non-enzymatic antioxidants and bacoside-A content in medicinal plant Bacopa monnieri L. Chemosphere 62:1340–1350
Sinha S, Saxena R, Singh S (2005) Chromium induced lipid peroxidation in the plants of Pistia stratiotes L.: role of antioxidants and antioxidant enzymes. Chemosphere 58:595–604
Suszcynsky EM, Shan JR (1995) Phytotoxicity and accumulation of mercury in tobacco subjected to different exposure routes. Environ Toxicol Chem 14:61–67
Taghinia Hejabi A, Basavarajappa HT, Qaid Saeed AM (2010) Heavy metal pollution in Kabini river sediments. Int J Environ Res 4(4):629–636
Venkatachalam P, Srivastava AK, Raghothama KG, Sahi SV (2009) Genes induced in response to mercury-ion-exposure in heavy metal hyperaccumulator Sesbania drummondii. Environ Sci Technol 43:843–850
Wang M, Zhou Q (2005) Single and joint toxicity of chlorimuron-ethyl, cadmium and copper acting on wheat Triticum aestivum. Ecotoxicol Environ Saf 60:169–175
Willekens H, Chamnongpol S, Davey M, Schraudner M, Langebartels C, Van Montagu M, Inzé D, Van Camp W (1997) Catalase is a sink for H2O2 and is indispensable for stress defense in C3 plants. Eur Mol Biol Org 16:4806–4816
Williams J, Kubelik AR, Livak KJ (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acid Res 18:6531–6535
Zavoda J, Cutright T, Szpak J, Fallon E (2001) Uptake, selectivity, and inhibition of hydroponic treatment of contaminants. J Environ Eng 127:502–508
Zhang J, Cui S, Kirkham MB (1995) Protoplasmic factors, antioxidant responses and chilling resistance in maize. Plant Physiol Biochem 33:567–575
Zhang FQ, Wang YS, Lou ZP, Dong JD (2007) Effect of heavy metal stress on antioxidative enzymes and lipid peroxidation in leaves and roots of two mangrove plant seedlings (Kandelia candel and Bruguiera gymnorrhiza). Chemosphere 67:44–50
Zhou ZS, Huang SQ, Guo K, Mehta SK, Zhang PC, Yang ZM (2007) Metabolic adaptation to mercury-induced oxidative stress in roots of Medicago sativa L. J Inorg Biochem 101:1–9
Acknowledgments
Authors gratefully acknowledge Mr. R. Manikandan, Department of Biotechnology, Periyar University, for his kind help rendered during statistical analysis.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Malar, S., Sahi, S.V., Favas, P.J.C. et al. Assessment of mercury heavy metal toxicity-induced physiochemical and molecular changes in Sesbania grandiflora L.. Int. J. Environ. Sci. Technol. 12, 3273–3282 (2015). https://doi.org/10.1007/s13762-014-0699-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13762-014-0699-4