Abstract
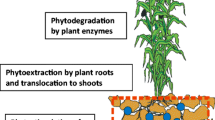
Synthetic organic compounds are hallmark of modern society. They are ubiquitous ranging from home, workplace to agriculture industry, which leads to their non-judicious dispensing into environment. Unfortunately most of them, especially polychlorinated biphenyls (PCBs), are deemed as persistent organic pollutants posing serious health risks to human. Hence, there is an alarming need of phasing out these chemicals and remediating contaminated sites in eco-friendly manner. Phytoremediation has emerged as a highly promising approach which capitalizes on plants and their associated microorganisms for removal of pollutants from targeted sites. Plant root exudations and secondary metabolites efficiently orchestrate selective recruitment of potential PCB-degrading microbial consortia within the rhizosphere and inside plant tissues. Structural analogy between organic contaminants and secondary plant metabolites (SPMEs) renders possible uptake and subsequent degradation of pollutants by microorganisms. Present review is focused on potential role of plant root exudates and SPMEs in structuring and orchestrating remediation of PCBs within rhizosphere and inside plant tissues. Also, recent developments in tools and techniques to study remediation of organic contaminants with special reference to PCBs are addressed.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
An increased anthropogenic activity due to industrial revolution is considered to be one of the major reasons behind indiscriminate dispensing of organic pollutants at massive rates in the environment. The advent of modern chemical industry has resulted in release of huge amounts of novel synthetic organic compounds, as industrial by-products, industrial solvents, pesticides, agrochemicals, pharmaceuticals, petroleum compounds, dioxins and furans, explosives, brominated flame retardants, polyaromatic hydrocarbons (PAHs), polychlorinated biphenyls (PCBs), etc. The Stockholm Convention on Persistent Organic Pollutants (POPs), under the auspices of the United Nations Environment Programme (UNEP), an international agreement enforced in 2004 (Stockholm convention 2004), enlisted twelve organic compounds as POPs. These twelve POPs include PCBs, nine chlorinated organic pesticides (aldrin, chlordane, dichlorodiphenyltrichloroethane (DDT), dieldrin, endrin, mirex, heptachlor, hexachlorobenzene and toxaphene), and dioxins and furans. PCB is a class of 209 chlorinated molecules called as congeners. Owing to their high chemical and thermal stability, commercial mixtures of PCBs were widely used in various industrial applications such as lubricants, plasticizers, dielectric fluids, hydraulic fluids in compressors and flame retardants (Borja et al. 2005). Although PCBs are no longer in use, but because of their extreme toxicity and persistence, they are still found as contaminants in the natural environments due to their past usage and continued to pose potential threats to human health. These compounds tend to resist biodegradation and persist in environment by attaching strongly to soil sediments and matrices, bioaccumulate in organisms especially at higher trophic level and finally enter food chains (Borja et al. 2005). The presence of ‘artificial’ groups such as chloro-, nitro- or sulfonate- in many synthetic chemicals makes them resistant to decomposition as they are not recognized by the degrading microbes. These compounds are collectively known as recalcitrant compounds. Various technologies and civil engineering methods such as excavation, land filling, thermal alkaline dechlorination or incineration (Campanella et al. 2002) appear to be expensive from both economic and environment stand point. Additionally, these methods may severely affect physical, chemical and biological properties of soil, thus, limiting their applications. These problems have interested researchers to look for the development of inventive remediation technologies for the cleanup of impacted sites and are therefore of paramount importance. Phytoremediation has emerged out as a newer, cleaner and promising approach which exploits plant and associated microorganism to remediate target site (Pulford and Watson 2003; Pilon-Smits 2005; Anyasi and Atagana 2011; Chen et al. 2013). It is a solar-energy-driven system that requires minimal site disturbance and maintenance, resulting in low-cost and high public acceptance due to great aesthetic value. Thus, last decade has gained tremendous momentum in the field of phytoremediation, and this approach has now entered into active field of development (Macek et al. 2000; Divya and Kumar 2011). Several plants including pine tree, alfalfa, flatpea, willow, canarygrass, deertongue, switchgrass, tall fescue, poplar, tobacco and mustard among others have been tested for their efficiencies to reduce PCBs in spiked soils (Ficko et al. 2010; Ionescu et al. 2009; Meggo et al. 2013). Low bioavailability in soil often results in limited utilization of PCBs by plants. Individual limitations of both plant and microbes for efficient removal of contaminants can be overcome by synergistic action of both plants and microbes. Microbe-assisted phytoremediation (rhizoremediation) with both native and introduced population (bioaugmentation) thus appear effective for enhanced degradation of PCBs (Wasi et al. 2013).
Plant metabolites fall in two main categories namely primary metabolites and secondary metabolites (SPMEs). Primary metabolites are involved in several essential phenomenons as growth, development and nutrition. Some examples of primary metabolites are, informational molecules as DNA and RNA, some energy-rich molecules as sucrose, starch, cellulose and pigments as chlorophyll. Secondary metabolites are generally not required by plants to survive but they have role in defense and protection against herbivory. SPMEs broadly fall into three structural categories, viz. terpenoids, phenolics, alkaloids, among others. Since antiquity SPMEs act as driving force for generating a vast spectrum of interactions among plant, insect and microorganisms ranging from symbiotic to antagonistic since antiquity. Moreover, SPMEs have significant roles in developing majority of degradative enzymes in nature due to immense diversity in their structures and analogy with typical organic contaminants.
Bacterial degradation of PCB
Microbial degradation of PCB is documented by several authors (Borja et al. 2005; Furukawa and Fujihara 2008). There are two major metabolic pathways: anaerobic dechlorination and aerobic biodegradation for PCB degradation. Often PCB congeners with four and higher chlorine atoms are degraded by anaerobic reductive dechlorination; it is an energy-yielding process in which PCBs act as electron acceptor for oxidation of organic carbon. Chlorine atoms are preferentially removed from the meta- and para-positions on the biphenyl structure, leaving lesser-chlorinated ortho-substituted congeners.
PCB dechlorination is mostly attributed to complex bacterial consortia. However, little is known about enzymes and molecular bases of metabolic pathways used in the process. Only a few bacterial species able to dechlorinate PCBs in pure culture have been identified, and their range of activity is limited to a few congeners. Microorganisms that reductively dechlorinate PCBs are widespread in contaminated sediments and involve species related to Dehalococcoides. Lower-chlorinated PCB congeners undergo co-metabolic aerobic oxidation mediated by dioxygenases, resulting in ring opening and potentially complete mineralization of the molecule. Members of several genera as Pseudomonas, Burkholderia, Comamonas, Rhodococcus, Achromobacter, Ochrobactrum, Stanotrophomonas and Bacillus are known to be potential PCB degraders. Aerobic biodegradation mediated by oxidative enzymes mainly depends upon the number and placement of chlorine atoms per molecule. PCB congeners with two to three chlorine atoms are easily degraded, while higher numbers of chlorine atoms are difficult to be degraded. Interestingly, Burkholderia xenovorans strain LB400, an extensively studied PCB degrader, was reported to metabolize a hexachlorobiphenyl congener (Field and Sierra-Alwarez 2008). Aerobic biodegradation of PCBs typically involves two clusters of genes, the first one is responsible for transformation into chlorobenzoates and chlorinated aliphatic acids (biphenyl upper pathway) and the second one is for further mineralization of chlorobenzoates and aliphatic acids (biphenyl lower pathway). The upper pathway, which is common in all described aerobic PCB degraders, involves seven genes grouped into one operon (biphenyl dioxygenase, bphA). A multi-component dioxygenase (bphA) initiates hydroxylation of two adjacent biphenyl carbons to form an arene cis-diol. In the second step, a cis-2, 3-dihydro-2, 3-dihydroxybiphenyl dehydrogenase (bphB) further oxidizes the biphenyl ring to produce a dihydroxychlorobiphenyl. In the third step, a second dioxygenase, 2, 3-dihydroxybiphenyl 1, 2-dioxygenase (bphC) opens the ring in ortho-meta-position. The final step of the upper pathway involves a hydrolase (bphD) that cleaves the resulting molecule into chlorobenzoate and 2-hydroxypenta-2, 4-dienoate (Fig. 1) (Furukawa and Fujihara 2008).
Rhizosphere: hot spot for pollutants degradation
Rhizosphere encompasses millimeters of soil surrounding plant root system (Hiltner 1904). It is the soil compartment where bacterial population is greatly influenced by rhizodeposits and plant age. On an average, 107–109 cfu (colony forming units) of rhizobacteria per gram of dry soil is found, approximately two- to threefolds higher when compared to bulk non-vegetated soil. Diverse array of plant root secretions (Flores et al. 1999) attract microorganisms toward roots where they establish themselves as colonizers. Hence, rhizosphere becomes hot spot for the complex interactions, because it is an active interface between living (microbes, insects, fungi, nematodes, etc.) and non-living (heavy metals, pesticides, chlorinated solvents, chemical signals, secondary plant metabolites, plant root exudates, etc.) components. These components further interact in intricate manner making this area more complex to understand (Bais et al. 2004). Plant root involved in various interactions in rhizosphere include root–root, root–insect and root–microbe interactions.
Several authors have confirmed positive effects of root exudates on increased microbial residency and activities in rhizosphere (Bais et al. 2004; Ionescu et al. 2009; Segura and Ramos 2012). These exudations are principal factors through which roots may regulate/alter the soil microbial community in their immediate vicinity, cope with herbivores, encourage beneficial symbioses, change the chemical and physical properties of the soil and inhibit growth of competing plant species (Nardi et al. 2000; Uhlik et al. 2012). Also, exudates direct some changes in transcriptional pattern of catabolic pathways in bacteria directing efficient utilization of pollutants. These exudates encompass nearly 5–21 % of all photosynthetically fixed carbon being transferred to the rhizosphere (Marschner 1995; Dennis et al. 2010). Soil microbes use these compounds as carbon and energy source (Leigh et al. 2002; Singer et al. 2004), thus supporting a diversity of microbial populations in the rhizosphere. Microbial consortia present in rhizosphere provide several benefits to plants including decrease in plant stress hormone level, chelation of key nutrient such as iron, protection against phytopathogen by secreting antibiotics and other bioactive compounds and degradation of contaminants (Glick 1995; Macek et al. 2000; Chaudhry et al. 2005; Liu et al. 2004). Variety of biochemical processes occurring in such a limited space of rhizosphere make it highly vulnerable to be manipulated to produce desired results, as improvement in crop production and active site for bioremediation takes shape.
Rhizoremediation is a microbe-assisted remediation approach, where root exudates and other plant-originated chemicals induce shifts in bacterial community and stimulate microbial populations to degrade organic pollutants (Glick 2003; Singer et al. 2004; Chaudhry et al. 2005). Successful rhizoremediation depends upon the extent of root colonization by degrading bacteria (Megharaj et al. 2011; Afzal et al. 2012; Masciandaro et al. 2013). Effective root colonization is attained by multitude of bacterial traits including chemotaxis, production of thiamine and biotin, synthesis of the O-antigen of lipopolysaccharide, amino acid synthesis and an efflux pump induced by isoflavonoids and motility (Capdevila et al. 2004; Compant et al. 2010).
These microbial associations play a central role in stepwise transformation of organic contaminants in the rhizosphere and within plants, and provide a milieu that supports genetic exchange and gene rearrangements. Contaminant-degrading bacteria can be isolated virtually from every gram of soil (Singer et al. 2004) with higher densities and activities in rhizosphere. Pollutant-degrading enzymes can also be found in plants, fungi, endophytic bacteria and root-colonizing bacteria, viz. peroxidases, dioxygenases, P450 mono-oxygenases, laccases, phosphatases, dehalogenases, nitrilases and nitroreductases (Susarla et al. 2002; Liu et al. 2004).
It is realized since ages that microorganisms have never remained in isolation; it is the continued influence of root exudates, cell debris and plants secondary metabolites (SPME’s) (Bais et al. 2004) that guides microbes (Challis and Hopwood 2003) to communicate and establish diverse relationship with one another. In some cases, production of root exudates may help improve PCB availability for degradation (Macek et al. 2000; White et al. 2006). Several investigators have already shown that specific plants may selectively foster PCB-degrading bacteria (Ryslava et al. 2003; Villacieros et al. 2003) (Table 1). Ionescu et al. 2009 screened four plants as Medicago sativa, Nicotiana tobacum, Salix caprea, Armoracia rusticana and Solanum nigrum to study the differential effect of plant species on microbial community. Authors have found that S. caprea and A. rusticana promoted the growth of potential PCB degraders better than N. tabacum and S. nigrum. In order to diagnose correlation between microbial count and PCB degradation, authors analyzed PCB content in soil both at the beginning and the end, i.e., after 6 months of plant cultivation of the experiment. They found that decline in PCB content in vegetated soil positively correlated with the numbers of culturable PCB degraders identified in the rhizosphere of individual plants; S. caprea and A. rusticana vegetated soil-mediated higher removal in PCB concentration, and this was further reflected in higher microbial counts when assessed for the increase in microbial activity. The rhizosphere of both species contained significantly higher number of PCB degraders. Characterization of bacterial isolates revealed a pronounced correlation between the plant species and bacterial genera recovered from rhizosphere of that plant, implying host specificity toward particular microbial population. For instance, S. nigrum promoted growth of Pseudomonas mendocina and Pseudomonas fluorescens. Sphingobacterium mizutae and Burkholderia cepacia were the possible PCB degraders found in the PCB soil vegetated by S. caprea, while Ochrobactrum anthropi and Agrobacterium radiobacter were found in other vegetated PCB soil samples and control. Also, isolates recovered from rhizosphere of S. caprea and A. rusticana exhibited broader range of PCB co-metabolism. This suggests that certain plant species are associated with high population sizes of PCB-metabolizing bacteria in root zone with broad congener specificities. In this study, A. rusticana and S. caprea were found to be good candidates for rhizoremediation of PCB-contaminated soil. Further, authors concluded that plants play an important role in direct and indirect removal of POPs. They accumulate and transform xenobiotics including PCBs (Rezek et al. 2007, 2008; Macci et al. 2012) and play an important role in providing nutrients and increase the bioavailability of pollutants for rhizospheric microorganisms or in soil close to the roots of plants. This corroborates the fact that plant species act differentially on microbial communities and certain plants significantly support degradation abilities of bacteria (Leigh et al. 2006). Alfalfa plantation treatment significantly lowers concentration of total extractable PCBs from soil (Chekol et al. 2004; Tu et al. 2011a, b). Authors have shown that there exists a significant effect of plant origin on degradation capacity of root zone bacteria. In second year, PCB degradation was much higher than first year in both treatment and control. Planting alfalfa significantly accelerates PCB biodegradation as 31.4 and 78.4 % of the initial PCBs were removed from planted treatments from the first and the second years, respectively, while only 12.3 and 31.4 % was removed in the unplanted control (Tu et al. 2011a, b). This increased degradation capability was due to increased microbial activities which was reflected in increased soil enzyme activities (dehydrogenase and FDA esterase). Since both enzymes exist in all microorganisms, they are known to give a measure of total viable microbial activities or act as bioindicators. Significantly, higher activity of both soil dehydrogenase and esterase was recorded in the treatment when compared to control. It was suggested that plants accelerate bioremediation by releasing secondary compounds including simple sugars, amino acids, flavonoids and aromatics that stimulate the growth of specific microbial communities in the soil (Macek et al. 2000) or possibly induce enzyme systems of the existing bacterial populations (Dudásová et al. 2012). Arabidopsis thaliana is known to contain flavanone as major component of root exudates that acts as biphenyl catabolic pathway inducer in a Rhodococcus erythropolis. Since structure of PCBs is analogues to flavonoids and coumarins, it is suggested that microbial populations residing in immediate vicinity of alfalfa plant root may be actively engaged in PCB degradation while utilizing these secreted compounds as carbon source.
Singer et al. (2003) studied the interactive effects of different treatments on the degradation of Aroclor 1242 in soil, including bioaugmentation with PCB-degrading bacteria, biostimulation with carvone, salicylic acid as inducers and sorbitan and trioleate as surfactants, and vegetation with Brassica nigra. These studies suggest that the increased depletion of the higher chlorinated PCB in the treated plants may be due to the specific deposition of aromatic-rich lignin and humic substances from plants. Table 1 gives the comprehensive data indicating various plant–microbe partnerships being exploited by various researchers to achieve PCB degradation.
Tu et al. (2011a, b) carried out experiments to study the potential role of PCB biodegradation by Sinorhizobium meliloti strain ATCC17519 in liquid cultures and soil microcosms. Authors found that S. meliloti could take 2, 4, 4′-TCB (PCB 28) as a sole carbon and energy source, without using biphenyl as the co-metabolic substrate. S. meliloti can transform more than 70 % of 2, 4, 4′-TCB, a highly efficient rhizobial strain (Damaj and Ahmad 1996). Further, S. meliloti could significantly enhance the depletion of all the twenty-one PCB congeners, especially to less chlorinated biphenyls. Other studies confirmed that inoculating alfalfa plants with R. meliloti further enhanced PCB removal from the rhizosphere soil, but microbial consortia working in tandem in the rhizosphere appear to be effective degraders (Macek et al. 2000; Kuiper et al. 2004; Chaudhry et al. 2005).
There is a vast battery of processes by which vegetation stimulates microbial activities which in turn enhance biodegradation of organic pollutants. The following tenets highlight comprehensive view on the degradative processes Van Aken et al. (2010):
-
1.
Organic compounds released by roots such as sugar, amino acids, and organic acids can act as electron donors to support aerobic co-metabolism or anaerobic dehalogenation of chlorinated compounds. In some instances, microbial aerobic metabolism will consume oxygen, resulting in anaerobic conditions favorable for dehalogenation of higher PCB (Chaudhry et al. 2005).
-
2.
Certain extracellular enzymes and inducers secreted by plants stimulate transformation of PCBs (Fletcher and Hegde 1995).
-
3.
Plant roots and root hair increase soil permeability and oxygen diffusion in the rhizosphere, which potentially enhances microbial oxidative transformation by oxygenases, key enzyme in degradation of biphenyl compounds (Chaudhry et al. 2005).
-
4.
Microbes usually experience difficulties in availing PCBs, being hydrophobic. Plant roots release organic acids and molecules that can act as surfactants; they help in mobilization PCBs, rendering them more susceptible to be absorbed inside plant tissues (Campanella et al. 2002).
Rhizoengineering
Plants have an active say in recruiting or exhibiting preferences toward a specialized group of population to colonize its rhizosphere. This microbial community is often referred as second genome of plant (Berendsen et al. 2012). Robust structural and functional diversities of plant root exudates and SPME’s is an eternal source to interdisciplinary researcher to explore the dynamic microcosm of the rhizosphere. Hence, it has been pronounced as an ideal site to modify microbial populations to suite various applications in the soil. The manipulation of plant root secretions and associated microbes to improve plant health and productivity is termed as rhizoengineering (O’Connell et al. 1996). Rhizoengineering exploits the strategies based on favouring the growth of the targeted microbes that possess the ability to metabolize exotic nutrients exuded by plants (Lugtenberg et al. 2001). A nutritional bias or favoritism is created, and population of bacteria can be further enriched which can preferentially utilize the given nutrients. Savka and Farrand (1997) were among the early reporter to achieve success in rhizoengineering based on favorably partitioning the exotic nutrient, opines, which were produced by the transgenic plants leading to improved and competitive growth of the metabolizing strains in comparison with the microbes unable to metabolize opines (opine concept). This strategy provides selective nutritional advantage to Agrobacterium and other selective bacteria considering genetic exchanges (Horizontal gene transfer). In yet another approach, various species of Pseudomonas selectively using salicylate as carbon source (Colbert et al. 1993) and detergents (Igepal Co-720 and dioctyl sulfosuccinate) from maize rhizosphere had been isolated. Flavanoids, most studied as chemicals involved in molecular cross-talk between plant roots and specific microorganisms, are known to induce transcription of nodulation genes in nitrogen-fixing rhizobia (Brencic and Winans 2005). Rhizobium sp. and Bradyrhizobium sp. transformed the flavonoids quercetin, daidzein and genistein through novel biochemical pathways in contrast to modes of degradation favored by other rhizobacteria (Toussaint et al. 2012). Flavanoids, naringenin and apigenin have been shown to support growth of PCB degradation by bacteria that are competent with respect to degradation of PCBs (Fletcher and Hegde 1995).Without a doubt, plant production of substrates that cause nutritional bias on the one hand and enzyme induction for the biodegradation of xenobiotic compounds on the other, offer intriguing and very attractive opportunities for rhizosphere manipulations (rhizoengineering) for enhancing rhizodegradation of xenobiotic contaminants beyond the inherent capabilities of the plant–microbe system. Further research may be directed to profiling of root exudates to identify compounds that may be used to create nutritional bias for enhancing rhizodegradation of persistent xenobiotic contaminants such as PCB (Narasimhan et al. 2003).
Structural analogy and organic contaminant degradation
The synergistic and antagonistic relationships between plants, microorganisms and insects (P-M-I) are responsible for the diversity of secondary plant metabolites. This P-M-I tritrophic interaction serves as one of the main driving forces of pollutant-degrading enzyme evolution (Singer et al. 2003, 2004).
How or why do microbes biodegrade synthetic recalcitrant compounds when these compounds are treated as fortuitous substrates by microbes? Perhaps, the continued exposure of pollutant analogues, albeit naturally created as dioxins (Meharg and Killham 2003), biphenyls (Liu et al. 2004) and volatile organic compounds, has helped microorganisms to develop pollutant-degrading abilities (Singer et al. 2003). It is hypothesized that most organic pollutants, although synthesized chemically, never appeared as aliens to microorganism since they bear tremendous structural analogy with plant-derived chemicals, hence stimulating microorganisms to degrade pollutants (Donnelly et al. 1994; Fletcher and Hegde 1995). Donnelly et al. (1994) were among the first investigators to study the link between plant-derived chemicals and pollutant remediation. The authors demonstrated that a range of flavanoids could support the growth of PCB-degrading microorganisms. Ralstonia eutrophus strain H850, B. cepacia LB400 and Corynebacterium sp. MB1 were studied in detail for congener depletion assay (Bedard et al. 1986), and it was found that naringin was proved to be the best growth substrate for H850 and supported its greatest metabolic activity on PCBs. Myricetin induced the greatest PCB degradation by LB400, which catabolized sixteen of the nineteen congeners tested. Strain MB1 degraded thirteen PCB congeners in the presence of coumarin in excess of the biphenyl controls. Another development was provided by Focht (1995) who suggested that plant terpenes, rather than biphenyl, may be the natural substrates for PCB catabolizing enzymes. Hernandez et al. (1997) further demonstrated that soils enriched with orange peel, ivy leaves, pine needles or eucalyptus leaves resulted in 105 times more biphenyl (unchlorinated PCB) utilizers (108 g−1) than their unsupplemented control (103 g−1), which suggested that terpenes might be natural substrates for biphenyl-utilizing bacteria (Hernandez et al. 1997). These authors showed that bacteria-utilizing cymene and limonene as sole carbon source transformed Aroclor 1242 (20–80 %) and (43–80 %), respectively, as compared to glucose utilizing bacteria. They postulated that slow-growing microorganisms (oligotrophs), relying on low concentrations and slowly delivered secondary plant metabolites, might be more effective in degrading PCBs. Gilbert and Crowley (1998) screened large number of plant (spearmint, pennyroyal, basil, barley, green bean, dill, avocado litter and garden compost) for the stimulation and degradation of 4-4′-dichlorobiphenyl by a known PCB-degrading bacterium, Arthrobacter sp., strain B1B. Analysis showed that carvone, a principal component of spearmint extract resulted in catabolic induction. Other terpenes as: p-cymene, isoprene, (S)-(+)-carvone, (R)-(−)-carvone, (S)-(−)-limonene, (R)-(+)-limonene, carvacrol, cumene, trans-cinnamic acid and thymol with structure similar to carvone were tested to induce PCB degradation(50 mg l−1), and it was found that all except cumene, trans-cinnamic acid and thymol enhanced 4,4′-dichlorobiphenyl metabolism, while p-cymene and isoprene accelerated catabolism in comparison with biphenyl (P < 0.05).
Isoprene, a non-aromatic compound lacking ring structure, was found to be one of the most effective inducer of PCB degradation. It was proposed that the relatively high antimicrobial activities of terpenes might induce a P450-like detoxification and fortuitous degradation of the PCBs. Cytochrome P450 enzymes are a large family of enzymes that have been shown to oxidize terpenes, such as camphor (P450 cam), as well as pollutants, such as polycyclic aromatic hydrocarbons (PAHs; e.g., naphthalene and pyrene, chlorinated phenols, and biphenyls. p-Cymene is widely studied SPME in pollutant-degrading experiments. Some authors argue that similarities in the amino acid sequence of enzymes catalyzing reactions in the p-cymene/cumate pathway and aromatic catabolic pathways are responsible for such activity. Its efficacy might also lie in (1) its structural similarity to several pollutants (e.g., toluene, xylene, ethylbenzene, biphenyl and chlorobenzene) and (2) a common evolutionary origin of the genes encoding the catabolic pathways (Eaton 1997).
Co-metabolite, inducer and surfactants: key to successful PCB degradation
PCB-degrading bacteria usually require biphenyl as co-metabolite, i.e., degradation of PCBs during growth on another substrate. PCBs transforming bacterial strains utilize the same sets of enzymes employed in the catabolism of biphenyl (Ahmed and Focht 1973). Biphenyl is often utilized as carbon source as well as an inducer of the necessary enzymes. Although biphenyl has been shown to be the best promoter of PCB degradation, its use in remediation is hindered by its own toxicity and low solubility (Luo et al. 2007).This problem can be solved by the use of several natural compounds, including terpenes, carvone, coumarin, etc., which have been shown to significantly induce PCB co-metabolism (Gilbert and Crowley 1998; Singer et al. 2000) (Table 2). The use of surfactants, which could increase the mass transfer and bioavailability of hydrophobic PCBs to microorganisms, has also been proposed for accelerated PCB degradation (Fava et al. 2003). Kang et al. (2010) successfully used microbial surfactant, sophorolipid for bioremediation of model hydrocarbons and crude oil in soil (Kang et al. 2010). These surfactants are known to increase desorption, apparent aqueous solubility and microbial bioavailability of hydrophobic organic carbons (HOCs).
Endophyte-assisted remediation: an edge over rhizosphere
Last decade has witnessed extensive and intensive research in the field of endophyte-mediated phytoremediation (Doty 2008). Endophytic bacteria colonize interior of plants and do not affect host plant adversely (Sessitsch et al. 2002). Endophytic bacteria are known to benefit plant through one or more properties such as nitrogen fixation, phytohormone production, inhibition of stress ethylene, induced systemic response and antagonistic activity (Parmeela and Johri 2004). Endophytes have edge over rhizospheric bacteria in terms of aiding phytoremediation as rhizospheric population is difficult to control, and competition between microbes often reduces the number of desired strains unless metabolism of pollutant is selective. Endophytes, in contrast, live in the internal tissues of the plant, and their populations seem to be selected or controlled by the plant. Therefore, the use of endophytes that naturally inhabit the plant would reduce the problem of competition. van Aken et al. (2004) have reported methyloptrophic bacteria from poplar tree (populus deltoids × populous nigra) that were capable of degrading the explosives trinitrotoluene (TNT), hexahydrotrinitrotriazine (RDX) and High Melting eXplosives (HMX); within 2 months, 60 % of RDX and HMX were mineralized to carbon dioxide. Volatile organic compounds (VOC’s) toluene and naphthalene are also reported to be degraded by endophytes isolated from hybrid poplar trees, (P. trichocarpa × P. deltoides cv. Hazendens and Hoogvorst) growing on a BTEX (benzene, toluene, ethylbenzene and xylene)-contaminated site in Belgium (Germaine et al. 2009). Out of 121 endophytic strains isolated from hybrid poplar trees, 34 were able to enhance phytoremediation. Germaine et al. (2006) successfully demonstrated the usefulness of bacterial endophytes Pseudomonas putida VM1450 in phytoremediation of 2, 4-dichlorophenoxyacetic acid. After inoculation of bacteria into pea and exposing plants to various levels of 2,4-D, it was found that plants showed higher potential of 2, 4-D removal from soil and sub-sequentially bearing no sign of 2, 4-D accumulation in aerial parts at any level of applied 2,4-D. While non-inoculated plants showed accumulation of 2,4-D corresponding to the increased levels of applied 2,4-D and accounted for 24–35 % of the total 2,4-D applied to the pots. P. putida VM1450 emerged as an efficient colonizer as well, and population sizes in the rhizosphere increased from an order of 105 with no selective pressure to between 106 and 107 when 13–54 mg of 2,4-D was added. The results showed that the presence of selective pressure, i.e., the introduction of 2, 4-D, had a positive effect on P. putida VM1450 populations within the rhizosphere, root and aerial tissues. This selective pressure effect was also reported by Jacobsen (1997) in a similar experiment where the author inoculated barley seeds with a Burkholderia strain, which possessed 2, 4-D degradation capability; this strain showed weak rhizosphere colonization ability in the absence of selective pressure, but when exposed to 10 or 100 mg kg−1 2, 4-D, the population grew to 107 cm−1 root.
Above-mentioned reports corroborate effect of selection pressure in recruiting desirable bacterial populations to efficient remediation of organic pollutants. Siciliano et al. (2001) elaborated that in response to routine exposure of plants to allelopathic compounds analogous to organic contaminants (Donnelly et al. 1994), there may be a plant response that stimulates microbial defense against a soil toxicant or toxin (Walton et al. 1994). The enrichment of catabolic genotypes in the root interior occurs in different plants in a variety of environments and in response to different contaminants. Hence, plants may be endowed with the ability to recruit bacteria that contain genotypes specific for toxic degradation into the rhizosphere and root interior, and this selection should be contaminant specific.
Endophytic microorganisms certainly have upper hand to improve xenobiotic remediation (Newman and Reynolds 2005) as genetic manipulation of degradation pathway in bacteria is easier than plants. In addition, monitoring the efficiency of remediation process can be done by quantitative gene expression of pollutant catabolic genes within the endophytic populations. Also larger population sizes can be easily reached in the absence of competition for substrate utilization. Most importantly, although plants often metabolize or sequester organics, they have a limited spectrum of organic compounds to be utilized as substrate. Also, with many substrates incomplete degradation results in accumulation of toxic compounds, this in turn may be detrimental to its sensitive organelles. Hence, they rely on associated microorganisms for an efficient degradation of organic compounds. Endophytes residing in immediate vicinity of these intermediate compounds can completely degrade toxic metabolites, which may be harmful to several organelles of plants, thereby eliminating the amount of toxic volatile compounds to be dispensed by plants through evapotranspiration.
Remediation with genetically modified Endophytes and plants
Last decade has witnessed global increase in research dedicated to improve abilities of plants and microbes to remove organic pollutants (Abhilash et al. 2009; James and Strand 2009). Several plant species have an inherent ability to metabolize a variety of environmental pollutants including insecticides, pesticides, herbicides, etc.
Genes from plants, microbes and animals have been used to enhance ability of plants to either remove or degrade contaminant. Both microbes and mammals are heterotrophic and possess several catabolic genes which can be used to complement the metabolic efficiencies of plants, e.g., mammalian genes encoding cytochrome P450s can lead to removal of pollutants and herbicides in tobacco. Unfortunately, plant cytochrome P450-mediated metabolism of PCBs results in toxic epoxides and trans-diol metabolites which cannot be further degraded, but bacterial biphenyl dioxygenase produces cis-diol intermediates which can be further mineralized completely (Sylvestre et al. 2009). French et al. (1999) succeed to introduce pentaerythritol tetranitrate (PETN) reductase gene into transgenic tobacco which resulted in increased tolerance to trinitroglycerin and TNT (French et al. 1999). Plant-associated endophytic bacteria can be genetically engineered to degrade toxic organic compounds to offer more potential than rhizospheric bacteria for reducing phytotoxicity (Barak et al. 2004). Endophytic bacteria can be isolated from host plants of interest (e.g., plants native to a geographical region) and genetically manipulated to contain degradation pathways or genes to degrade target contaminants before being reinoculated back into the host plant for bioremediation purposes. Germaine et al. (2009) constructed a naphthalene-degrading endophytic strain P. putida VM1441 (pNAH7), colonizing efficiently both within the rhizosphere and interior root tissues. P. putida VM1441 (pNAH7) inoculation into plant resulted in higher rates of seed germination, plant transpiration and protection of the host plant from phytotoxic effects of naphthalene. Further, higher naphthalene degradation rates (40 %) were achieved when compared with uninoculated plants in artificially spiked soil. In yet another ground-breaking study, Barac et al. introduced pTOM, a toluene-degrading plasmid into B. cepacia L.S.2.4, a natural endophyte of yellow lupine plant from B. cepacia G4. After successful inoculation of engineered strain into yellow lupine seed (surface sterile), it efficiently degraded toluene with marked decrease in phytotoxicity and 50–70 % decrease in evapotranspiration through leaves. As an extension to this study, Taghavi et al. (2005) showed that horizontal gene transfer of pTOM to a number of endophytes in planta was possible, promoting more efficient degradation of toluene in poplar plants. Horizontal gene transfer (HGT) in planta is likely to be widespread, as studies in pea with Pseudomonas endophytes harboring the plasmids Pwwo and pNAH7 also showed high rates of transfer into a range of autochthonous endophytes (Ryan et al. 2007). Phytoremediation of herbicides has been enhanced by using transgenic plants expressing GST (Karavangeli et al. 2005) or cytochrome P450 genes (Kawahigashi et al. 2006).
However, despite several successful attempts to achieve enhanced remediation rates with engineered plants and microbes, this technology is still in its infancy and declined public acceptance. The reports at field-level applications (testing) of transgenic plants are scarce. Major obstacle is biosafety concern, because the effects of genetically modified organisms (GMOs) are still not fully understood. It is thought that they may alter structure of indigenous microbial community. Soil microorganisms play significant roles in several organic and inorganic matters, and alteration in structure and functionality of such organisms may pose adverse effects on ecology and productivity. Several studies report that microbial communities are severely affected by engineered plants in soil (Bruce et al. 2007; LeBlanc et al. 2007; Lee et al. 2011) while others advocate that either the detrimental effects posed are too minor or they are statistically non-significant (Schmalenberger and Tebbe 2002; Dunfield and Germida 2003; Kapur et al. 2010). Most of these studies are based on non-sequencing-based methods as community-level physiological profiles (CLPPs), fatty acid methyl esters (FAME), denaturation gradient gel electrophoresis (DGGE) and terminal restriction length polymorphism (T-RFLP) analysis. However, these methods have limited resolution capabilities to detect minor changes in community structure as only a small portion of genetic material is probed. Thus, a thorough and well-planned study utilizing next-generation sequencing (NGS) technology is required in order to have a complete idea of microbial communities dwelling with GM (genetically modified) plants. Another major concern is loss in species diversity of soil microorganisms resulting in loss of some important processes occurring naturally in soil. Outbreak of new diseases and occurrence of unexpected results are also some of the challenges faced by scientists till date. However, it is realized that both the risks and benefits cannot be generalized. It depends on crop to crop and very much on temporal and spatial events Wolenbarger and Phifer (2000).
Recent tools and techniques to study organic contaminant degradation
Rhizosphere is a continually evolving system offering multifacets of exciting fronts to researchers. Advancements of highly sophisticated techniques in molecular biology have brought a new dawn to study organic pollutants degradation under in situ conditions. With the advent of techniques bypassing need of culturing bacteria under laboratory conditions (Spring et al. 1992), ground-breaking results were achieved. Not only community dynamics but functional community dynamics of both culturable and culture independent bacteria using DNA-based stable isotope probing (DNA-SIP) technique can be diagnosed. This approach utilizes specific consumption of a given substrate carrying a 13C signature and can be associated with the small subunit ribosomal RNA molecules of the microbes that consume it (Whiteley et al. 2007).
In this technique, DNA is used as the labeled biomarker (DNA-SIP), subsequently coupled with harnessing the superior phylogenetic resolution of the small subunit (SSU) ribosomal RNA gene (Radajewski et al. 2000). Uhlik et al. (2012) in an interesting study investigated naringenin, caffeic acid and limonene-induced shifts in bacterial community composition with their degradative capabilities on long-term PCB-contaminated soil. They integrated pyrosequencing of 16s rRNA gene–gene tag-encoded amplicons and DNA-SIP to analyze and identify populations actively involved in 4-chlorobiphenyl catabolism. Authors concluded that out of three PSMs used, application of naringin showed efficient degradation when compared to control, and Hydrgenophaga, Terrimonas, Paucimonas and Pseudorhodoferax were among the most dominant genera identified. The second major improvement is the use of the SSU rRNA molecule itself (RNA-SIP) (Manefield et al. 2002). Radajewski et al. (2000) enhanced the phylogenetic resolution of SIP by demonstrating that stable isotope labeled DNA could be isolated from mixed microbial communities; since isotopic enrichment increases buoyant density of DNA, subsequent density centrifugation in CsCl gradients can be used to separate ‘heavy’ (labeled) from ‘natural’ (unlabeled) DNA; finally, 16S rDNA clone libraries are constructed from ‘heavy’ DNA and sequenced to obtain the identity of organisms assimilating the 13 C labeled substrate used.
Casavant et al. (2003) used a gfp-labeled P. fluorescens A506 to detect low concentrations of toluene (0.2 mm) and trichloroethylene in the rhizosphere and observed an increased gfp expression in the root-colonizing biosensor population when the plant rhizosphere was exposed to toluene. Interestingly, they noted the presence of natural inducers as 14 % more induced cells were observed in uncontaminated rhizosphere soil than bulk soil. It was further confirmed that promoter is induced by variety of alkyl-substituted benzene derivatives and branched alkenes (Singer et al. 2003).
Rhizospheric metabolomics, a similar application to the ‘field application vector’ approach, was applied by Narasimhan et al. (2003). These workers demonstrated that during PCB degradation in rhizosphere of Arabidopsis, phenylpropanoid-utilizing microbes are more competitive and are able to grow at least 100-fold better than their auxotrophic mutants on roots of plants that are able to synthesize or overproduce phenylpropanoids, such as flavonoids. This finding further supports the fact that plants remain the ultimate driving force behind recruitment of microbes within the rhizosphere.
Biosensors are useful tools when carrying out environmental risk assessments on polluted sites or for monitoring the efficacy of a remediation strategy, due to their ability to detect only the biologically relevant (bio-available) fraction of the contaminant. Liu et al. (2004) constructed three biosensor strains; P. fluorescens F113rifgfp, P. fluorescens F113rifPCBgfp and P. fluorescens F113L::1180gfp designed to detect the bi-availability and biodegradation of PCBs. As the F113rifPCBgfp and F113L::1180gfp strains degraded PCBs, they produced chlorinated benzoate end products which in turn induced the expression of gfp. Therefore, these strains could actively report on their own degradation of PCBs. Use of biosensors strains describes the application to monitor PCB biodegradation in different PCB-contaminated soils and sediments. This approach can be exploited in both soil bioremediation and plant rhizoremediation. Some authors suggested that by immobilizing the biosensor cells, ease of detection, accuracy and reproducibility could be improved. There exists a positive correlation between the PCB levels within the samples and the percentage of fluorescing biosensors cells when immobilized PCB-degrading biosensors (F113L::1180gfp and F113rifPCBgfp) were introduced into PCB-contaminated soil and sludge. Greater number of fluorescent cells is a clear cut indication of greater bioavailability of PCBs and their degradation by the GM biosensors which in turn indicated that the soil/sludge posed a greater risk to human health. Demnerova et al. (2005) carried out PCB degradation experiments using three plants species. Their results supported previous existing plethora of data that the number of natural PCB degraders on roots was at least ten times higher than that in bulk soil. Gfp expressing biosensor cells were easily detectable, and they were in higher densities in soils planted with pea, indicating increased PCB degradation activity compared with unplanted soil. Authors stated that this study demonstrated that biosensors could be used at two important fronts, firstly, to evaluate a specific contaminated site for bioremediation potential, and secondly, to monitor PCB degradation in real PCB-contaminated soil.
Further, use of real-time PCR increases the probability of accurately determining population sizes of PCB-degrading microbes. Biosensor cell numbers detected by qRT-PCR method is tenfold higher than the numbers detected using the plate counting method. The high sensitivity of real-time PCR which could detect culturable, non-culturable inoculants together with dead inoculants (Cubero and Graham 2005). It was also documented previously by Wang et al. (2004) that using a qRT-PCR method, a tenfold higher level of the engineered P. putida could be detected compared to the plate counting method, during 2-chlorobenzoate degradation in soil.
Genetic fingerprinting methods as denaturation gradient gel electrophoresis (DGGE) and terminal restriction fragmentation length polymorphism (T-RFLP) are proven to be useful in determining spatial and temporal changes in bacterial communities present (Anderson et al. 2010).
Conclusion
This Host–microbe interaction specificity is a center of muse and manipulations for researchers since eternity. Plants are endowed with enormous potential and functions since they evolved on earth. Xenobiotic compounds degradation can be understood as bipartite phenomenon. Plants on one hand uptake, accumulate and partially metabolize organic contaminants, and on the another hand, compounds produced by plants allow survival of microorganisms even in poor soils, serve as carbon and energy source, increase soil aeration and can even induce the degradation pathways of vast majority of xenobiotics in them. They influence the composition of consortia of microorganisms in a manner that positively favors them, in soil surrounding roots and within rhizosphere. Thus, the choice of proper plant species becomes crucial for addressing effective cleaning of polluted sites. Again, to achieve a substantial degradation, the soil conditions have to be manipulated artificially such as addition of various amendments and surfactants, etc. to increase bioavailability and mobility to ensure optimal composition and population of microbial consortia by selecting right plants and nutrients. An optimal cocktail of plant root exudates, secondary metabolites, surfactants and carbon source amendments coupled with appropriate localization of potential degraders present as indigenous microbiota can be constructed order to achieve complete removal of contaminated sites.
References
Abhilash PC, Jamil S, Singh N (2009) Transgenic plants for enhanced biodegradation and phytoremediation of organic xenobiotics. Biotechnol Adv 27:474–488
Afzal M, Yousaf S, Reichenauer TG, Sessitsch A (2012) The inoculation method affects colonization and performance of bacterial inoculant strains in the phytoremediation of soil contaminated with diesel oil. Int J Phytoremediation 14:35–47
Ahmed M, Focht DD (1973) Degradation of polychlorinated biphenyls by two species of Achromobacter. Can J Microbiol 19:47–52
Anderson SA, Northcote PT, Page MJ (2010) Spatial and temporal variation of the bacterial community in different chemotypes of the New Zealand marine sponge Mycale hentscheli. FEMS Microbiol Ecol. doi:10.1111/j.1574-6941.2010.00869.x
Anyasi RO, Atagana HI (2011) Biological remediation of polychlorinated biphenyls (PCBs) in the environment by microorganisms and plants. Afr J Biotechnol 10(82):18916–18938. doi:10.5897/AJB10.557
Bais HP, Park SW, Weir TL, Callaway RM, Vivanco JM (2004) How plants communicate using the underground information superhighway. Trends Plant Sci 9:26–32
Barak T, Taghavi S, Borremans B, Provoost A, Oeyen L, Colpaert JV, Vangronsveld J, van der Lelie D (2004) Engineered endophytic bacteria improve phytoremediation of water-soluble, volatile, organic pollutants. Nat Biotechnol 22:583–588
Bedard DL, Unterman R, Bopp LH, Brennan MJ, Haberl ML, Johnson C (1986) Rapid assay for screening and characterizing microorganisms for the ability to degrade polychlorinated biphenyls. Appl Environ Microbiol 51:761–768
Berendsen RL, Pieterse CMJ, Bakker PAHM (2012) The rhizosphere microbiome and plant health. Trends Plant Sci. doi:10.1016/j.tplants.2012.04.001
Borja J, Taleon DM, Auresenia J, Gallardo S (2005) Polychlorinated biphenyls and their biodegradation. Process Biochem 40:1999–2013
Brencic A, Winans SC (2005) Detection of and response to signals involved in host–microbe interactions by plant-associated bacteria. Microbiol Mol Biol Rev 69:155–194
Bruce NC et al (2007) Impact of transgenic tobacco on trinitrotoluene (TNT) contaminates soil community. Environ Sci Technol 41:5854–5861
Campanella BF, Bock C, Schroder P (2002) Phytoremediation to increase the degradation of PCBs and PCDD/Fs—Potential and limitations. Environ Sci Pollut Res 9:73–85
Capdevila S, Martınez-Granero FM, Sanchez-Contreras M, Rivilla R, Martın M (2004) Analysis of Pseudomonas fluorescens F113 genes implicated in flagellar filament synthesis and their role in competitive root colonization. Microbiology 150:3889–3897
Casavant NC, Thompson D, Beattie GA, Phillips GJ, Halverson LJ (2003) Use of a site-specific recombination based biosensors for detecting bioavailable toluene and related compounds in roots. Environ Microbiol 5:238–249
Challis GL, Hopwood DA (2003) Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc Natl Acad Sci 100:14555–14561
Chaudhry Q, Blom-Zandstra M, Gupta S, Joner EJ (2005) Utilizing the synergy between plants and rhizosphere microorganisms to enhance breakdown of organic pollutants in the environment. Environ Sci Pollut Res 12:34–48
Chekol T, Vough LR, Chaney RL (2004) Phytoremediation of polychlorinated biphenyl-contaminated soils: the rhizosphere effect. Environ Int 30:799–804
Chen J, Xu QX, Su Y, Shi ZQ, Han FX (2013) Phytoremediation of organic polluted soil. J Bioremediation Biodegradation 4:132
Colbert SF, Schroth MN, Weinhold AR, Hendson M (1993) Enhancement of population densities of Pseudomonas putida PpG7 in agricultural ecosystem by selective feeding with carbon source salicylate. Appl Envrion Microbiol 59(7):2064–2070
Compant S, Clément C, Sessitsch A (2010) Plant growth-promoting bacteria in the rhizo- and endo-sphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol Biochem 42:669–678
Cubero J, Graham JH (2005) Quantitative real-time polymerase chain reaction for bacterial enumeration and allelic discrimination to differentiate xanthomonas strains on citrus. Bacteriology 95:1333–1340
Damaj M, Ahmad D (1996) Biodegradation of polychlorinated biphenyls by rhizobia: a novel finding. Biochem Biophys Res Commun 218:908–915
Demnerova K, Mackova M, Spevakova V, Beranova K, Kochankova L, Lovecka P et al (2005) Two approaches to biological decontamination of ground-water and soil polluted by aromatics-characterization of microbial populations. Int Microbiol 8:205–211
Dennis PG, Miller AJ, Hirsch PR (2010) Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol Ecol 72:313–327
Divya B, Kumar DM (2011) Plant–microbe interaction with enhanced bioremediation. Res J Biotechnol 4(6):72–79
Donnelly PK, Hegde RS, Fletcher JS (1994) Growth of PCB-degrading bacteria on compounds from photosynthetic plants. Chemosphere 28:981–988
Doty SL (2008) Enhancing phytoremediation through the use of transgenics and endophytes. New Phytol 179:318–333
Dudásová H, Lukácová L, Murínová S, Dercová K (2012) Effects of plant terpenes on biodegradation of polychlorinated biphenyls (PCBs). Int Biodeterior Biodegradation 69:23–27
Dunfield KF, Germida JJ (2003) Seasonal changes in the rhizosphere microbial communities associated with the field-grown genetically modified canola (Bassica napus). Appl Environ Microbiol 69:7310–7318
Eaton RW (1997) p-Cymene catabolic pathway Pseudomonas putida F1: cloning and characterization of DNA encoding conversion of p-cymene to p-cumate. J Bacteriol 179(10):3171–3180
Fava F, Bertin L, Fedi S, Zannoni D (2003) Methyl-beta-cyclodextrin-enhanced solubilization and aerobic biodegradation of polychlorinated biphenyls in two aged-contaminated soils. Biotechnol Bioeng 81:381–390
Ficko SA, Rutter A, Zeeb BA (2010) Potential for phytoextraction of PCBs from contaminated soils using weeds. Sci Total Environ 408:3469–3476
Field JA, Sierra-Alwarez R (2008) Microbial transformation and degradation of polychlorinated biphenyls. Environ Pollut 1551:1–12
Fletcher JS, Hegde RS (1995) Release of phenols by perennial plant roots and their potential importance in bioremediation. Chemosphere 31:3009–3016
Flores HE, Vivanco JM, Loyola-Vargas VM (1999) “Radicle” biochemistry: the biology of root-specific metabolism. Trends Plant Sci 4:220–226
Focht DD (1995) Strategies for the improvement of aerobic metabolism of polychlorinated-biphenyls. Curr Opin Biotechnol 6:341–346
French CE, Rosser SJ, Davis GJ, Nicklin S, Bruce NC (1999) Biodegradation of explosives by transgenic plants expressing pentaerythritol tetranitrate reductase. Nat Biotechnol 17:491–494
Furukawa K, Fujihara H (2008) Microbial degradation of polychlorinated biphenyls: biochemical and molecular features. J Biosci Bioeng 105:433–449
Germaine KJ, Liu X, Cabellos GG, Hogan JP, Ryan D, Dowling DN (2006) Bacterial endophyte-enhanced phytoremediation of the organochlorine herbicide 2,4-dichlorophenoxyacetic acid. FEMS Microbiol Ecol 57:302–310
Germaine KJ, Keogh E, Ryan D, Dowling DN (2009) Bacterial endophyte-mediated naphthalene phytoprotection and phytoremediation. FEMS Microbiol Lett 296:226–234
Gilbert ES, Crowley DE (1997) Plants compound that induce polychlorinated biphenyls degradation by Arthrobacter sp. strain B1B. Appl. Environ Microbiol 63:1933-1938
Gilbert ES, Crowley DE (1998) Repeated application of carvone-induced bacteria to enhance biodegradation of polychlorinated biphenyls in soil. Appl Microbiol Biotechnol 50:489–494
Glick BR (1995) The enhancement of plant growth by free-living bacteria. Can J Microbiol 41:109–117
Glick BR (2003) Phytoremediation: synergistic use of plants and bacteria to clean up the environment. Biotechnol Adv 21:383–393
Hernandez BS, Koh S-C, Chiral M, Focht DD (1997) Terpene utilizing isolates and their relevance to enhanced biodegradation of polychlorinated biphenyls in soil. Biodegradation 8:153–158
Hiltner L (1904) Über neue Erfahrungen und Probleme auf dem Gebiet der Bodenbakteriologie und unter besonderer Berücksichtigung der Gründüngung undBrache. Arbeiten der Deutschen Landwirtschafts-Gesellschaft 98:59–78
Ionescu M, Beranova K, Dudkova V, Kochankova L, Demnerova K, Macek T, Mackova M (2009) Isolation and characterization of different plant associated bacteria and their potential to degrade polychlorinated biphenyls. Int Biodeterior Biodegradation 63:667–672
Jacobsen CS (1997) Plant protection and rhizosphere colonisation of barley by seed inoculated, herbicide degrading Burkholderia (Pseudomonas) cepacia DB01 (pR0101) in 2, 4-D contaminated soil. Plant Soil 189:139–144
James CA, Strand SE (2009) Phytoremediation of small organic contaminants using transgenic plants. Curr Opin Biotechnol 20:237–241
Kang S-W, Kim Y-B, Shim J-D, Kim E-K (2010) Enhanced biodegradation of hydrocarbons in soil by microbial biosurfactant, Sophorolipid. Appl Biochem Biotechnol 160:780–790
Kapur M, Bhatia R, Pandey G, Pandey JD, Jain RK (2010) A case study for assessment of microbial community dynamics in genetically modified Bt cotton crop fields. Curr Microbiol 61:118–124
Karavangeli M, Labrou NE, Clonis YD, Tsaftaris A (2005) Development of transgenic tobacco plants overexpressing maize glutathione transferase I for chloroacetanilide herbicides phytoremediation. Biomol Eng 22:121–128
Kawahigashi H, Hirose S, Ohkawa H, Ohkawa Y (2006) Transgenic rice plants expressing human P450 genes involved in xenobiotic metabolism for phytoremediation. J Agric Food Chem 54:2985–2991
Kim BH, Oh ET, So JS, Ahn Y, Koh SC (2003) Plant terpenes-induced expression of multiple aromatic ring hydroxylation oxygenase genes in Rhodococcus sp. strain T104. J Microbiol 41:349-352
Kuiper I, Lagendijk EL, Bloemberg GV, Lugtenberg BJ (2004) Rhizoremediation: a beneficial plant–microbe interaction. Mol Plant Microbe Interact 17:6–15
LeBlanc PM, Hamelin RC, Filion M (2007) Alteration of soil rhizosphere communities following genetic transformation of white spruce. Appl Environ Microbiol 73:4128–4138
Lee YE, Yang SH, Bae TW, Kang HG, Lim PO, Lee HY (2011) Effects of field-grown genetically modified zoysia grass on bacterial community structure. J Microbiol Biotechnol 21:333–340
Leigh MB, Fletcher JS, Fu X, Schmitz FJ (2002) Root turnover: an important source of microbial substrates in rhizosphere remediation of recalcitrant contaminants. Environ Sci Technol 36:1579–1583
Leigh MB, Prouzová P, Macková M, Macek T, Nagle DP, Fletcher JS (2006) Polychlorinated biphenyl (PCB)-degrading bacteria associated with trees in a PCB-contaminated site. Appl Environ Microbiol 72:2331–2342
Liu B, Beuerle T, Klundt T, Beerhues L (2004) Biphenyl synthase from yeast-extract-treated cell cultures of Sorbus aucuparia. Planta 218:492–496
Lugtenberg BJ, Dekkers L, Bloemberg GV (2001) Molecular determinants of rhizosphere colonization by Pseudomonas. Annu Rev Phytopathol 39:461–490
Luo WS, D’Angelo EM, Coyne MS (2007) Plant secondary metabolites, biphenyl and hydroxypropyl-beta-cyclodextrin effects on aerobic polychlorinated biphenyl removal and microbial community structure in soils. Soil Biol Biochem 39:735–743
Macci C, Doni S, Peruzzi E, Ceccanti B, Masciandaro G (2012) Bioremediation of polluted soil through the combined application of plants, earthworms and organic matter. J Environ Monit 14:2710–2717
Macek T, Mackova M, Kas J (2000) Exploitation of plants for the removal of organics in environmental remediation. Biotechnol Adv 18:23–34
Maeda M Chung SY, Song E, Kudo T (1995) Multiple genes encoding 2,3- dihydroxybiphenyl 1,2- dioxygenase in the gram positive polychlorinated biphenyls-degrading bacterium Rhodococcus erythropolis TA421, isolated from a termite system. Appl Environ Microbiol 61:549-555
Manefield M, Whiteley AS, Griffiths RI, Bailey MJ (2002) RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl Environ Microbiol 68:5367–5373
Marschner H (1995) Mineral nutrition of higher plants. Academic Press, London
Masciandaro G, Macci C, Peruzzi E, Ceccanti B, Doni S (2013) Organic matter–microorganism–plant in soil bioremediation: a synergic approach. Rev Environ Sci Biotechnol. doi:10.1007/s11157-013-9313-3
Meggo RE, Schnoor JL, Hu D (2013) Dechlorination of PCBs in the rhizosphere of switchgrass and poplar. Eniviron Pollut 178:312–321
Megharaj M, Ramakrishnan B, Venkateswarlu K, Sethunathan N, Naidu R (2011) Bioremediation approaches for organic pollutants: a critical perspective. Environ Int 37:1362–1375
Meharg AA, Killham K (2003) A pre-industrial source of dioxins and furans. Nature 421:909–910
Narasimhan K, Basheer C, Bajic VB, Swarup S (2003) Enhancement of plant-microbe interactions using a rhizosphere metabolimics-driven approach and its application in the removal of polychlorinated biphenyls. Plant Physiol 132:146–153
Nardi S, Concheri G, Pizzeghello D, Sturaro A, Rella R, Parvoli G (2000) Soil organic matter mobilization by root exudates. Chemosphere 5:653–658
Newman L, Reynolds C (2005) Bacteria and phytoremediation: new uses for endophytic bacteria in plants. Trends Biotechnol 23:6–8
O’Connell KP, Goodman RM, Handelsman J (1996) Engineering the rhizosphere: expressing a bias. Trends Biotechnol 14:83–88
Oh ET, Koh S-C, Kim E, Ahn Y-H, So J-S (2003) Plant terpenes enhance survivability of polychlorinated biphenyls (PCBs) degrading Pseudomonas Pseudoalcaligenes KF707 labeled with gfp in microcosms contaminated with PCB. J Microbiol Biotechnol 13:463-468
Park Y-I, So J-S, Koh S-C (1999) Induction by carvone of the polychlorinated biphenyls (PCBs)-degradative pathway in Alcaligenes eutropa H850 and its molecular monitoring. J Microbiol Biotechnol 9:804-810
Parmeela, Johri BN (2004) Phylogenetic analysis bacterial endophytes showing anatagonism against Rhizoctonia solani. Curr Sci 8(5):687–692
Pilon-Smits E (2005) Phytoremediation. Annu Rev Plant Biol 56:15–39
Pulford ID, Watson C (2003) Phytoremediation of heavy-metal-contaminated land by trees: a review. Environ Int 29:529–540
Radajewski S, Ineson P, Parekh NR, Murrell JC (2000) Stable-isotope probing as a tool in microbial ecology. Nature 403:646–649
Rezek J, Macek T, Mackova M, Triska J (2007) Plant metabolites of polychlorinated biphenyls in hairy root culture of black nightshade Solanum nigrum SNC-9O. Chemosphere 69:1221–1227
Rezek J, Macek T, Mackova M, Triska J, Ruzickova K (2008) Hydroxy-PCBs, methoxy-PCBs and hydroxy-methoxy-PCBs: metabolites of polychlorinated biphenyls formed in vitro by tobacco cells. Environ Sci Technol 42:5746–5751
Ryan RP, Ryan D, Dowling DN (2007) Plant protection by the recombinant, root-colonising Pseudomonas fluorescens F113rifPCB strain expressing arsenic resistance: improving rhizoremediation. Lett Appl Microbiol 45(6):668–674
Ryslava E, Krejcik Z, Macek T, Novakova H, Demnerova K, Mackova M (2003) Study of PCB degradation in real contaminated soil. Fresenius Environ Bull 12:296–301
Savka MA, Farrand SK (1997) Modification of rhizobacterial populations by engineering bacterium utilization of novel plant-produced resource. Nat Biotechnol 15:363–368
Schell MA (1985) Transcriptional control of the nah and sal hydrocarbon- degradation genes from plasmid NAH7. J Bacteriol 26:2049-2057
Schmalenberger A, Tebbe CC (2002) Bacterial community composition in the rhizosphere of a transgenic, herbicide-resistant maize (Zea mays) and comparison on its non-transgenic cltivar Bosphore. FEMS Microbiol 40:29–37
Segura A, Ramos JL (2012) Plant-bacteria interactions in the removal of pollutants. Curr Opion Biotechnol 24:1–7
Sessitsch A, Reiter B, Pfeifer U, Wilhelm E (2002) Cultivation-independent population analysis of bacterial endophytes in three potato varieties based on eubacterial and Actinomycetes-specific PCR of 16s rRNA genes. FEMS Microbiol Ecol 39:3–32
Siciliano S, Fortin N, Himoc A et al (2001) Selection of specific endophytic bacterial genotypes by plants in response to soil contamination. Appl Environ Microbiol 67:2469–2475
Singer AC, Gilbert ES, Luepromchai E, Crowley DE (2000) Bioremediation of polychlorinated biphenyl-contaminated soil using carvone and surfactant-grown bacteria. Appl Microbiol Biotechnol 54:838–843
Singer AC, Crowley DE, Thompson IP (2003) Secondary plant metabolites in phytoremediation and biotransformation. Trends Biotechnol 21:123–130
Singer AC, Thompson IP, Bailey MJ (2004) The tritrophic trinity: a source of pollutant-degrading enzymes and its implications for phytoremediation. Curr Opin Microbiol 7(3):239–244
Siciliano SD, Germida JJ (1998) Mechanisms of phytoremediation: biochemical and ecological interactions between plants and bacteria. Environ Rev 6:65–79
Spring S, Amann R, Ludwig W, Schleifer K-H, Petersen N (1992) Phylogenetic diversity and identification of nonculturable magnetotactic bacteria. Syst Appl Microbiol 15(1):116–122
Susarla S, Medina VF, McCutcheon SC (2002) Phytoremediation: an ecological solution to organic chemical contamination. Ecol Eng 18:647–658
Sylvestre M, Macek T, Mackova M (2009) Transgenic plants to improve rhizoremediation of polychlorinated biphenyls (PCBs). Curr Opin Biotechnol 20:242–247
Taghavi S, Barac T, Greenberg B et al (2005) Horizontal gene transfer to endogenous endophytic bacteria from poplar improves phytoremediation and development of poplar trees. Appl Environ Microbiol 75:748–757
Tandlich R, Brezna B, Dercova K (2001) The effect of terpenes on the biodegradation of polychlorinated biphenyls by Pseudomonas stutzeri. Chemosphere 44:1547–1555
Tang, J, Wang, R, Niu X, Wang M, Zhou Q (2010a) Characterization on the rhizoremediation of petroleum contaminated soil as affected by different influencing factors. Biogeosci Discuss 7:4665–4688
Toussaint JP, Pham T, Barriault D, Sylvestre M (2012) Plant exudates promote PCB degradation by a Rhodococcal rhizobacteria. Appl Microbiol Biotechnol 95:1589–1603
Tu C, Teng Y, Luo Y, Li X, Sun X, Li Z, Liu W, Christies P (2011a) Potential for biodegradation of polychlorinated biphenyls (PCBs) by Sinorhizobium meliloti. J Hazard Mater 186:1438–1444
Tu C, Ying T, Yongming L, Xianghui S, Shaopo D, Zhengao L, Wuxing L, Zhihong X (2011b) PCB removal, soil enzyme activities, and microbial community structures during the phytoremediation by alfalfa in field soils. J Soils Sediments 11:649–656
Uhlik O, Musilova L, Hroudova RM, Vlcek C, Koubek J, Holeckova M, Mackova M, Macek T (2012) Plant secondary metabolites-induced shifts in bacterial community structure and degradative ability in contaminated soil. Appl Micobiol Biotechnol. doi:10.1007/s00253-012-4627-6
Van Aken B, Yoon JM, Just CL, Schnoor JL (2004) Metabolism and mineralization of hexahydro-1,3,5-trinitro-1,3,5-triazine inside poplar tissues (populus deltoids X populous nigra DN-34). Environ Sci Technol 38:4572–4579
Van Aken B, Correa PA, Schnoor JL (2010) Phytoremediation of polychlorinated biphenyls: new trends and promises. Enivron Sci Technol 44:2767–2776
Villacieros M, Power B, Sanchez-Contreras M, Lloret J, Oruezabal RI, Martin M (2003) Colonization behaviour of Pseudomonas fluorescens and Sinorhizobium meliloti in the alfalfa (Medicago sativa) rhizosphere. Plant Soil 251:47–54
Walton BT, Hoylman AM, Perez MM, Anderson TA, Johnson TR, Guthrie EA, Christman RF (1994) Rhizosphere microbial communities as a plant defense against toxic substances in soils. In: Anderson TA, Coats JR (eds) Bioremediation through rhizosphere technology. ACS Symposium Series 563. American Chemical Society, Washington, DC, pp. 82–92
Wang G, Gentry T, Grass G, Josephson K, Rensing C, Pepper IL (2004) Real-time PCR quantification of a green fluorescent protein-labelled, genetically engineered Pseudomonas putida strain during 2-chlorobenzoate degradation in soil. FEMS Microbiol Lett 233:307–314
Wasi S, Tabrez S, Ahmad M (2013) Use of Pseudomonas spp. for the bioremediation of environmental pollutants: a review. Environ Monit Assess. doi:10.1007/s10661-013-3163-x
White JC, Ross DW, Gent MPN, Eitzer BD, Mattina MI (2006) Effect of mycorrhizal fungi on the phytoextraction of weathered p, p-DDE by Cucurbita pepo. J Hazard Mater 137:1750–1757
Whiteley AS, Thomson B, Lueders T, Manefield M (2007) RNA-stable isotope probing. Nat Protoc 2:838–844
Wolenbarger LL, Phifer PR (2000) The ecological risks and benefits of genetically engineered plants. Science 290:2088–2093
Acknowledgments
Corresponding author thanks Department of Science and Technology, India, for financial support as DST, women scientist, DST No. SR/WOS-A/LS-275/2011 (G). Authors thank Prof. B.N. Johri for critical reading, valuable comments and suggestions on this manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jha, P., Panwar, J. & Jha, P.N. Secondary plant metabolites and root exudates: guiding tools for polychlorinated biphenyl biodegradation. Int. J. Environ. Sci. Technol. 12, 789–802 (2015). https://doi.org/10.1007/s13762-014-0515-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13762-014-0515-1