Abstract
In this paper, we analyze the problem of data clustering in domains where discrete and continuous variables coexist. We propose the use of hybrid Bayesian networks with naïve Bayes structure and hidden class variable. The model integrates discrete and continuous features, by representing the conditional distributions as mixtures of truncated exponentials (MTEs). The number of classes is determined through an iterative procedure based on a variation of the data augmentation algorithm. The new model is compared with an EM-based clustering algorithm where each class model is a product of conditionally independent probability distributions and the number of clusters is decided by using a cross-validation scheme. Experiments carried out over real-world and synthetic data sets show that the proposal is competitive with state-of-the-art methods. Even though the methodology introduced in this manuscript is based on the use of MTEs, it can be easily instantiated to other similar models, like the Mixtures of Polynomials or the mixtures of truncated basis functions in general.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Unsupervised classification, cluster analysis or clustering [1, 14] are different ways of referring to the task of searching for decompositions or partitions of a data set into groups in such a way that the objects in one group are similar to each other, but at the same time as different as possible to the objects in other groups.
According to the result they produce, clustering methods can be classified as hard or soft. Hard clustering algorithms (like \(k\) means and hierarchical clustering) yield rules that assign each individual to a single group, therefore producing classes with sharp bounds. On the other hand, soft clustering (like probabilistic clustering) allows the possibility that an individual can belong to more than one cluster, according to a probability distribution.
In this paper, we focus on probabilistic clustering, and from this point of view the data clustering problem can be regarded as the estimation of a joint probability distribution for the variables in the database. Our starting point is the preliminary study carried out by Gámez et al. [12], where the MTE (Mixture of Truncated Exponential) model [23] was applied to the data clustering problem. The MTE model has been shown as a competitive alternative to the use of Gaussian models in different tasks like inference [17, 28], supervised classification [9], regression [8, 24] and learning [27], but the applicability to the clustering paradigm is still limited to the above-mentioned work.
In this paper, we introduce new algorithms that improve the performance of the seminal model proposed by Gámez et al. [12] and make it competitive with respect to the state-of-the-art methods. The new contributions of this paper are the use of a procedure for translating a Gaussian distribution into an ad hoc MTE approximation in the presence of normality, the possibility of re-assigning values of the hidden variable depending on the likelihood values in each iteration of the data augmentation process, the random selection of the cluster to split when increasing the number clusters and the use of the BIC score [30] to measure the quality of the intermediate models.
The paper is structured as follows: first, Sects. 2 and 3 give, respectively, some preliminaries about probabilistic clustering and mixtures of truncated exponentials. Section 4 describes the original model proposed by Gámez et al. [12] to obtain a clustering from data by using mixtures of truncated exponentials to model continuous variables and presents the new algorithms developed. Experiments over several datasets are described in Sect. 5, and in Sect. 6 we summarize the methodology proposed, discuss about some improvements to the method and outline future work.
2 Probabilistic model-based clustering
The usual way of modeling data clustering in a probabilistic approach is to add a hidden random variable to the data set, i.e., a variable whose value is missing in all the records. The hidden variable, usually referred to as the class variable, reflects the cluster membership for every individual in the data set.
Probabilistic model-based clustering is defined as a mixture of models (see e.g. [5]), where the states of the hidden class variable correspond to the components of the mixture (the number of clusters). A commonly used approach consist of modeling the discrete variables with a multinomial distribution while a Gaussian distribution is used to model the continuous variables. In this way, probabilistic clustering can be regarded as learning from unlabeled data, and usually the EM algorithm [4] is used to carry out the learning task.
In this paper, we focus on representing the joint distribution of the variables in the database using the so-called naïve Bayes structure [22], which assumes that all the variables are independent when the value of the class is known. Graphically, the naïve Bayes model can be represented as a Bayesian network [26], where each node represents a random variable and contains a marginal distribution for such variable if the node has no parents, or a conditional distribution given its parents otherwise. Figure 1 shows an example of a Bayesian network representing a naïve Bayes model. The joint distribution is factorized as
Assuming a naïve Bayes model, the clustering problem involves the determination of the number of clusters (\(k\)), i.e., the number of possible values for variable \(C\) in Fig. 1, and estimate the parameters of the distributions in Eq. (1). This is achieved by using the EM (expectation-maximization) algorithm [4]. The EM algorithm works iteratively by carrying out the following two-steps at each iteration:
-
Expectation Estimate the distribution of the hidden variable \(C\), conditioned on the current setting of the parameter vector \(\theta ^k\) (i.e., the parameters needed to specify the non-hidden variable distributions).
-
Maximization Taking into account the new distribution of \(C\), obtain by maximum-likelihood estimation a new set of parameters \(\theta ^{k+1}\) from the observed data.
The algorithm starts from a given initial starting point (e.g. random parameters for \(C\) or \(\theta \)) and under fairly general conditions it is guaranteed to converge to (at least) a local maximum of the log-likelihood function.
Iterative approaches have been described in the literature [2] in order to discover also the number of clusters (components of the mixture). The idea is to start with \(k=2\) and use EM to fit a model, then the algorithm tries with \(k=3, 4, \ldots \) as long as the log-likelihood improves by adding a new component to the mixture.
An important issue in probabilistic clustering is how to evaluate an obtained model. A common approach is to use the log-likelihood of the used dataset (\(\mathbf {D}\)) given the model to score a given clustering, since it is a probabilistic description of the dataset:
The above measure was used in Gámez et al. [12]. In this paper, we propose to use the BIC score instead which includes a penalty factor that takes into account the size of the model.
3 Mixtures of truncated exponentials
The MTE model [23] was introduced as an alternative to conditional Gaussian models, Lauritzen and Wermuth [20] and can also be seen as a generalization of discretization models. It is defined by its corresponding potential and density as follows.
Definition 1
(MTE potential) Let \(\mathbf {X}\) be a mixed \(n\)-dimensional random vector. Let \(\mathbf {Y}\!=\! (Y_1,\ldots ,Y_c)^{\mathsf {T}}\) and \(\mathbf {Z} \!=\! (Z_1,\ldots ,Z_d)^{\mathsf {T}}\) be the continuous and discrete parts of \(\mathbf {X}\), respectively, with \(c+d=n\). We say that a function \(f:\Omega _{\mathbf {X}}\mapsto \mathbb {R}_0^+\) is a Mixture of Truncated Exponentials potential (MTE potential) if one of the next conditions holds:
-
(i)
\(\mathbf {Z}=\emptyset \) and \(f\) can be written as
$$\begin{aligned}&f(\mathbf {x}) = f(\mathbf {y}) = a_0 + \sum _{i=1}^m a_i \mathrm {e}^{\mathbf {b}_{i} ^{\mathsf {T}}\mathbf {y}} \end{aligned}$$(3)for all \(\mathbf {y}\in \Omega _{\mathbf {Y}}\), where \(a_i \in \mathbb {R}\) and \(\mathbf {b}_i \in \mathbb {R}^{c}\), \(i=1,\ldots ,m\).
-
(ii)
\(\mathbf {Z}=\emptyset \) and there is a partition \(D_1,\ldots ,D_k\) of \(\Omega _{\mathbf {Y}}\) into hypercubes such that \(f\) is defined as
$$\begin{aligned}&f(\mathbf {x}) = f(\mathbf {y}) = f_i(\mathbf {y}) \quad \mathrm {if}~~ \mathbf {y}\in D_i, \end{aligned}$$where each \(f_i\), \(i=1,\ldots ,k\) can be written in the form of Eq. (3).
-
(iii)
\(\mathbf {Z}\ne \emptyset \) and for each fixed value \(\mathbf {z}\in \Omega _{\mathbf {Z}}\), \(f_{\mathbf {z}}(\mathbf {y})=f(\mathbf {z},\mathbf {y})\) can be defined as in ii.
Example 1
The function \(f\) defined as
is an MTE potential since all of its parts are MTE potentials.
An MTE potential \(f\) is an MTE density if:
A conditional MTE density can be specified by dividing the domain of the conditioning variables and specifying an MTE density for the conditioned variable for each configuration of splits of the conditioning variables.
Example 2
Consider two continuous variables \(X\) and \(Y\). A possible conditional MTE density for \(X\) given \(Y\) is the following:
Since MTEs are defined into hypercubes, they admit a tree-structured representation in a natural way. Moral et al. [23] proposed a data structure to represent MTE potentials, which is specially appropriate for this kind of conditional densities: the so-called mixed probability trees or mixed trees for short. The formal definition is as follows:
Definition 2
(Mixed tree) We say that a tree \(\mathcal{T}\) is a mixed tree if it meets the following conditions:
-
(i)
Every internal node represents a random variable (discrete or continuous).
-
(ii)
Every arc outgoing from a continuous variable \(Y\) is labeled with an interval of values of \(Y\), so that the domain of \(Y\) is the union of the intervals corresponding to the arcs emanating from \(Y\).
-
(iii)
Every discrete variable has a number of outgoing arcs equal to its number of states.
-
(iv)
Each leaf node contains an MTE potential defined on variables in the path from the root to that leaf.
Mixed trees can represent MTE potentials defined by parts. Each entire branch in the tree determines one hypercube where the potential is defined, and the function stored in the leaf of a branch is the definition of the potential on it.
Example 3
Consider the following MTE potential, defined for a discrete variable (\(Z_1\)) and two continuous variables (\(Y_1\) and \(Y_2\)).
A representation of this potential by means of a mixed probability tree is displayed in Fig. 2.
A marginal MTE density \(f(y)\), for a continuous variable \(Y\), can be learnt from a sample as follows [27]:
-
1.
Select the number \(k\) of intervals to split the domain.
-
2.
Select the number \(m\) of exponential terms on each interval.
-
3.
A Gaussian kernel density is fitted to the data.
-
4.
The domain of \(Y\) is split into \(k\) pieces, according to changes in concavity/convexity or increase/decrease in the kernel density.
-
5.
In each piece, an MTE potential with \(m\) exponential terms is fitted to the kernel density by iterative least squares estimation.
-
6.
Update the MTE coefficients to integrate up to one.
When learning an MTE density \(f(z)\) from a sample, if \(Z\) is discrete, the procedure is:
1. For each state \(z_i\) of \(Z\), compute the maximum likelihood estimation of \(f(z_i)\), which is \(\displaystyle \frac{\#z_i}{\text {sample size}}\).
A conditional MTE density can be specified by dividing the domain of the conditioning variables and specifying an MTE density for the conditioned variable for each configuration of splits of the conditioning variables [27]. In this paper, the conditional MTE densities needed are \(f(x\mid c)\), where \(C\) is the class variable, which is discrete, and \(X\) is either a discrete or continuous variable, so the potential is composed by an MTE potential defined for \(X\), for each state of the conditioning variable \(C\).
In general, a conditional MTE density \(f(x\mid y)\) with one conditioning discrete variable \(Y\), with states \(y_1, \ldots , y_s\), is learnt from a database as follows:
-
1.
Divide the database into \(s\) sets, according to the states of variable \(Y\).
-
2.
For each set, learn a marginal density for \(X\), as described above, with \(k\) and \(m\) constant.
4 Probabilistic clustering using MTEs
4.1 Algorithm
In the clustering algorithm proposed here, the conditional distribution for each variable given the class is modeled by an MTE density. In the MTE framework, the domain of the variables is split into pieces and in each resulting interval, an MTE potential is fitted to the data. In this work we will use the so-called \(five\)-parameter MTE, which means that in each split there are five parameters to be estimated from data:
The choice of the \(five\)-parameter MTE is motivated by its low complexity and high fitting power [3]. The number of splits of the domain of each variable has been set to four, again according to the results in the cited reference.
We start the algorithm with two clusters with the same probability, i.e., the hidden class variable has two states and the marginal probability of each state is \(1/2\). The conditional distribution for each feature variable given each one of the two possible values of the class is initially the same, and is computed as the marginal MTE density estimated from the training database according to the estimation algorithm described in Sect. 3. The steps for learning an initial model are formally shown in Algorithm 3 (see Appendix).
The initial model is refined using the data augmentation method [13] in order to obtain the best model with two clusters in terms of likelihood:
-
1.
First, the training database is divided into two parts, training set and test set, one for estimating the models in each iteration and the other one for validating them.
-
2.
For each record in the training and test sets, the distribution of the class variable is computed conditional on the values of the features in the record.
-
3.
According to the obtained distribution, a value for the class variable is simulated and inserted in the corresponding cell of the database.
-
4.
When a value for the class variable has been simulated for all the records, the (conditional) distributions are re-estimated using the method described in Sect. 3, using as sample the training set.
-
5.
The log-likelihood of the new model is computed from the test set. If it is higher than the log-likelihood of the initial model, the process is repeated. Otherwise, the process is stopped, returning the best model found for two clusters.
This data augmentation procedure has been partially modified in our proposal (see Algorithm 4). After imputing a class value after the simulation step, we try to speed up the convergence of the algorithm by updating the simulated class values by those ones that cause the best likelihood of the record with respect to the current dataset (see Algorithm 5). For the sake of efficiency, this replacement is only carried out for those records with the lowest likelihood. The fraction of updated records is in fact an input parameter of the algorithm.
Another issue we have modified from the data augmentation proposal by Gilks et al. [13] that we have adopted in this paper is related to the criterion for repeating the iterative process (see Algorithm 4). We have included a BIC score instead of a single likelihood measure in order to penalize the number of clusters in the model (note that this DA algorithm is not only applied to refine the model with two clusters, but also with more than two). More details about this BIC score are explained in Sect. 4.3.
After obtaining the best model for two clusters, the algorithm continues increasing the number of clusters and repeating the procedure described above for three clusters, and so on. Algorithm 2 shows the steps for including a new cluster in the model (i.e., a new state for the hidden class variable). This is based on the ideas for component splitting and parameter reusing proposed by Karciauskas [15].
The idea is to split one existing cluster into two by setting the same probability distribution for both, and ensuring that all the potentials integrate up to 1. The key point here is to find the optimal cluster to split. In that sense, a random subset of possible splits is checked, and the best partition in terms of likelihood is selected. The percentage of clusters to be checked is also an input parameter of the algorithm that will be analyzed in the experiment section. The process of adding clusters to the model is repeated while the BIC score of the model, computed from the test database, is increased.
At this point, we can formulate the main clustering algorithm (Algorithm 1). It receives as argument a database with variables \(X_1,\ldots ,X_n\) and returns a naïve Bayes model with variables \(X_1,\ldots ,X_n,C\), which can be used to predict the class value of any object characterized by features \(X_1,\ldots ,X_n\).
4.2 Conversion from Gaussians to MTEs
The procedure for learning an MTE potential from data presented in Sect. 3 does not take into account the underlying distribution of the data, i.e., when an MTE density is learnt from the corresponding data in a hypercube, the parameters are estimated by least squares fitting previously a kernel density on that hypercube.
The improvement proposed in this section relies on studying the normality of the data in each cluster when a conditional MTE density is being estimated. Thus, if a normality test is satisfied, we replace the learning approach [27] proposed so far by a procedure to approximate a Gaussian distribution with parameters \(\mu \) and \(\sigma \) by an MTE density [7]. Notice that the normality assumption can be satisfied in one cluster and not in another of the same conditional MTE potential. From a computational point of view, carrying out a normality test and constructing an MTE density from a Gaussian density if faster than directly learning an MTE potential from data. Beyond the increase in efficiency, under the normality assumption, an MTE potential obtained by directly learning from data is in general less accurate than another one induced by estimating the mean and variance from the data and translating the corresponding Gaussian to an MTE.
Methods for obtaining an MTE approximation of a Gaussian distribution are described in previous works [3, 18]. Common to both approaches is the fact that the split points used in the domain of the MTE approximation depend on the mean and variance of the original Gaussian distribution. Consider a Gaussian variable \(X \sim \mathcal{N}(\mu ,\sigma ^2)\). To obtain an MTE representation of the Gaussian distribution, we follow the procedure by Langseth et al. [18] in which no split points are used, except to define the boundary. A 6-term MTE density has been selected to take part of the approximation because it represents a tradeoff between complexity and accuracy.
Notice that this approximation is only positive within the interval \([\mu -2.5\sigma ,\mu +2.5\sigma ]\), and it actually integrates up to 0.9876 in that region, which means that there is a probability mass of \(0.0124\) outside this interval. In order to avoid problems with \(0\) probabilities, we add tails covering the remaining probability mass. More precisely, we define the normalization constant
and include the tail
for the interval above \(x=\mu +2.5\sigma \) in the MTE specification. Similarly, a tail is also included for the interval below \(x=\mu -2.5\sigma \). The transformation rule from Gaussian to MTE therefore becomes
where the parameters \(a_i\) and \(b_j\) are:
Figure 3 shows the MTE approximation to a standard Gaussian density using the 3 pieces specified in Eq. (5).
4.3 The BIC score for validating models
In probabilistic clustering, if the parameters are learnt by maximum likelihood, increasing the number of clusters in the model yields a model with higher likelihood, but also with higher complexity. In our proposal, it is not always the case, since the parameters in the MTE densities are estimated by least squares instead of maximum likelihood [18]. In order to reach a tradeoff between accuracy and complexity, and to avoid overfitting, we proposed the use of the BIC score instead of the log-likelihood when validating the inclusion of a new cluster in the model. The BIC score particularized to the MTE framework is computed as:
where \(\text {dim}(f)\) is the number of free parameters in the MTE model computed as:
where \(p\) is the number of MTE potentials in the model, \(q\) is the number of leaves in the mixed tree representing the \(j\)-th potential and \(m_{jk}\) is the number of exponential terms in the \(k\)th leaf of the \(j\)th potential.
5 Experimental evaluation
In order to analyze the performance of the proposed clustering algorithm, we have implemented the algorithm in the Elvira environment [6] and conducted a set of experiments on the databases described in Table 1. Out of the 8 databases, 7 refer to real-world problems, while the other one (net15) is a randomly generated network with joint MTE distribution [27]. Some of the databases are oriented to supervised classification, so that we have modified them by removing all the values in the column corresponding to the class variable.
Using the data sets described above, the following experimental procedure was carried out:
-
As a benchmark, we used the EM algorithm implemented in the Weka data mining suite [35]. This is a classical implementation of the EM algorithm in which discrete and continuous variables are allowed as input attributes. A naïve Bayes graphical structure is used, where each class model is a product of conditionally independent probability distributions. In this particular implementation, continuous attributes are modeled by means of Gaussian distributions and the number of clusters can be specified by the user or directly detected by the EM algorithm by using cross-validation [33]. In such case, the number of clusters is initially set to 1, the EM algorithm is performed by using a tenfold cross-validation and the log-likelihood (averaged for the tenfolds) is computed. Then, the number of clusters is increased by one and the same process is carried out. If the log-likelihood with the new number of clusters has increased with respect to the previous one, the execution continues, otherwise the clustering obtained in the previous iteration is returned.
-
From each database, we extracted a holdout set (20 % of the instances) used to assess the quality of the final models. The remaining 80 % of the instances is given as input for the data augmentation and EM algorithms.
-
The experiments in Elvira have been repeated five times due to the stochastic nature of the algorithm and the average log-likelihood per case has been reported. Since the number of clusters discovered by the algorithm may be different in each run, the median of the five numbers has been considered.
-
Both implementations (Elvira and Weka) include iteratively a new cluster in the model when the quality of the model improves. In the case of Elvira, models up to ten clusters are checked even if the BIC score does not improve with respect to the previous models. Thus, above ten, the algorithm includes a cluster only if the BIC score grows with respect to the best model found so far.
Several experiments haven been carried out in order to analyze the behavior of the different issues proposed in Sect. 4.
5.1 Experiment 1: conversion from Gaussians to MTEs
This experiment is aimed at analyzing the impact of including in the algorithm the conversion from Gaussians to MTEs proposed in Sect. 4.2, with respect to the previous MTE learning setting [27]. To do this, two versions of the Algorithm 1 were performed, one excluding the conversion, and the other, including it. In both cases, \(100~\%\) of clusters were checked when adding a new cluster in the model, and \(50~\%\) of hidden values were checked after data augmentation method. The results are shown in Tables 2 and 3, respectively. Note that, besides obtaining the optimum number of clusters for each approach (Elvira and Weka) and its associated likelihood, there is an extra column in order to compare the performance of both algorithms over the same number of clusters.
The log-likelihood of the best algorithm for each dataset is shown in italic type. Comparing both approaches, it is clear that there is an improvement in terms of likelihood in the MTE approach when considering the conversion in those clusters where the data normality is satisfied. In fact, without applying the conversion (Table 2) the MTE method outperforms the EM algorithm in 3 out of 8 datasets, whilst introducing the conversion (Table 3), the result is 4 out of 8. Moreover, the log-likehood of all the datasets in which we loose have been also increased with the conversion, specially abalone from a value of \(6.79\) to \(9.78\).
A rough analysis of the results suggests a similar performance for both approaches. However, it must be pointed out that the number of clusters obtained for the best models generated by the EM is significantly higher, indicating that it needs to include more clusters to get competitive results. In practice, it may happen that a model with many clusters, all of them with low probability, is not as useful as a model with fewer clusters but with higher probabilities, even if the latter one has lower likelihood.
The results obtained for the different datasets are highly influenced by the distribution of their variables. The higher the number of Gaussian variables, the better the behavior obtained by the EM approach, since it is specially designed for Gaussian distributions. To justify this statement, the number of Gaussian variables for each dataset and the best algorithm in terms of likelihood are shown in Table 4. We can see that the EM algorithm in Weka only takes advantage when the underlying data distribution is mostly Gaussian, whilst the MTE-based algorithm obtains better results in datasets with any other underlying data distribution.
After this analysis, we consider the MTE-based clustering algorithm a promising method. It must be taken into account that the EM algorithm implemented in Weka is very optimized and sophisticated, while the MTE-based method presented here is a first version in which many aspects are still to be improved. Even in these conditions, our method is competitive in some problems. Furthermore, datasets which are away from normality (as net15) are more appropriate for applying the MTE-based algorithm rather than EM.
5.2 Experiment 2: varying input parameters
In all the experiments mentioned from now on, the conversion from Gaussians to MTEs described in Sect. 4.2 was used. Experiment 2 was designed for exploring the behavior of the algorithm when changing the two input parameters explained in Sect. 4.1, namely pStates (\(\%\) of states checked when including a new cluster in the model) and pInstances (\(\%\) of instances checked to update its hidden value to that one causing the best likelihood of the record). The different scenarios for both parameters are described in Table 5.
The results for the three settings are shown in Tables 3, 6 and 7, respectively. Note that the results for setting 1 have already been showed in the experiment 1.
The results for the three different scenarios of input parameters indicate that the best performance of the MTE-based algorithm takes place under the setting 1, in which the percentage of both parameters is the highest (100 and \(50~\%\), respectively). In this case, our proposal outperforms the EM algorithm in 4 out of 8 datasets. However, as the percentage of the input parameters is reduced, the results become worse (3 out of 8 under setting 2, and only in 2 under setting 3).
5.3 Experiment 3: BIC score
This experiment studies the effect of including the BIC score when learning the MTE model. Table 8 shows the log-likelihood per case of the MTE versus the EM approach. A priori, it is sensible to expect that the number of clusters with respect to those obtained in Table 3 should be reduced, since the BIC score penalizes models with more parameters. However, as shown in Table 8 the number of clusters is not always reduced (see discussion in Sect. 4.3).
5.4 Statistical tests
Several tests have been carried out to detect differences between the various proposals discussed throughout the paper. Table 9 shows the result of applying the Wilcoxon test to the likelihood results for all the combinations of settings of our algorithm and the EM approach. On the other hand, Table 10 shows the results of the same statistical test after comparing, for every table, the best likelihood obtained by the MTE model (column 3 in Tables 2, 3, 6, 7, 8) versus the likelihood obtained by the EM forcing the number of clusters obtained by the MTE (column 4 in Tables 2, 3, 6, 7, 8), and the other way around for column 6 and 7. The results in Tables 9 and 10 indicate the absence of statistically significant differences between the various settings studied.
There is also no difference between the number of clusters obtained before and after including the BIC score in learning the model (\(p\) value \(=0.907\)).
6 Discussion and concluding remarks
In this paper, we present the MTE framework as an alternative to the Gaussian model to deal with unsupervised classification. Based on the work by Gámez et al. [12], we develop a new methodology able to perform clustering over datasets with both discrete and continuous variables without assuming any underlying distribution. The model has been tested over 8 databases, 7 of them corresponding to real-world problems commonly used for benchmarks. As the statistical tests show, the model, when comparing the results to the refined EM algorithm implemented in the Weka data mining suite, is competitive, since no significative differences have been found. However, we can see that, in the absence of Gaussian variables in the data, our proposed method attains better results than the EM algorithm (except for abalone dataset). Furthermore, in the presence of Gaussian variables, the results have improved when using the conversion. The algorithm we have used in this paper can still be improved. For instance, during the model learning we have used a train-test approach for estimating the parameters and validating the intermediate models obtained. However, the implementation of the EM algorithm in Weka uses cross-validation. We plan to include tenfold cross-validation in a forthcoming version of the algorithm.
A natural extension of this methodology is the application of more sophisticated structures beyond the naïve Bayes, such as the TAN [11], kdB [29] or the AODE [34] models. The greatest advantage of using the MTE framework within these models is that it is allowed to define a conditional distribution over a discrete variable with continuous parents, which is forbidden in the case of the Gaussian model. However, as the complexity increases, new methods to efficiently implement the ideas presented in this paper on these more elaborated models need to be devised.
An important feature of the technique presented in this paper is that it can be directly applied using frameworks related to MTEs, like the Mixtures of Polynomials (MOPs) [32] and more generally, Mixtures of Truncated Basis Functions (MoTBFs) [19]. This can lead to improvements in inference efficiency, especially using MoTBFs, as they can provide accurate estimations with no need to split the domain of the densities [16]. However, we decided to use just MTEs for the sake of computational efficiency. In this paper, we have considered the inclusion of only two exponential terms into a fixed number of subintervals of the range of every variable, whilst MoTBFs in general result in an unbounded number of terms without splitting the domain of the variable. We have resorted to the original estimation algorithm in [27], because it requires fewer iterations for learning the parameters than the general MoTBF algorithm [16].
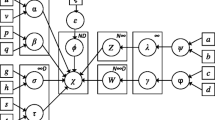
The algorithm proposed in this paper can be applied not only to clustering problems, but also to construct an algorithm for approximate probability propagation in hybrid Bayesian networks. This idea was firstly proposed by Lowd and Domingos [21], and is based on the simplicity of probability propagation in naïve Bayes structures. A naïve Bayes model with discrete class variable, actually represents the conditional distribution of each variable to the rest of variables as a mixture of marginal distributions, being the number of components in the mixture equal to the number of states of the class variable. So, instead of constructing a general Bayesian network from a database, if the goal is probability propagation, this approach can be competitive. The idea is specially interesting for the MTE model, since probability propagation has a high complexity for this model.
References
Anderberg, M.: Cluster Analysis for Applications. Academic Press, New York (1973)
Cheeseman, P., Stutz, J.: Bayesian classification (AUTOCLASS): theory and results. In: Fayyad, U.M., Piatetsky-Shapiro, G., Smyth, P., Uthurusamy, R. (eds.) Advances in Knowledge Discovery and Data Mining, pp. 153–180. AAAI Press/MIT Press, Cambridge (1996)
Cobb, B.R., Shenoy, P.P., Rumí, R.: Approximating probability density functions with mixtures of truncated exponentials. Stat. Comput. 16, 293–308 (2006)
Dempster, A.P., Laird, N.M., Rubin, D.B.: Maximum Likelihood from Incomplete Data via the EM Algorithm. J. R. Stat. Soc. B 39, 1–38 (1977)
Duda, R.O., Hart, P.E., Stork, D.G.: Pattern Classification. Wiley Interscience, New York (2001)
Elvira-Consortium: Elvira: an environment for creating and using probabilistic graphical models. In: Proceedings of the First European Workshop on Probabilistic Graphical Models, pp. 222–230 (2002). http://leo.ugr.es/elvira
Fernández, A., Langseth, H., Nielsen, T.D., Salmerón, A.: Parameter learning in MTE networks using incomplete data. In: Proceedings of the Fifth European Workshop on Probabilistic, Graphical Models (PGM’10), pp. 137–145 (2010)
Fernández, A., Nielsen, J.D., Salmerón, A.: Learning bayesian networks for regression from incomplete databases. Int. J. Uncertain. Fuzziness Knowl. Based Syst. 18, 69–86 (2010)
Flores, J., Gámez, J.A., Martínez, A.M., Salmerón, A.: Mixture of truncated exponentials in supervised classification: case study for naive Bayes and averaged one-dependence estimators. In: Proceedings of the 11th International Conference on Intelligent Systems Design and Applications, pp. 593–598 (2011)
Flores, M., Gámez, J., Mateo, J.: Mining the ESROM: a study of breeding value classification in Manchego sheep by means of attribute selection and construction. Comput. Electron. Agric. 60(2), 167–177 (2008)
Friedman, N., Geiger, D., Goldszmidt, M.: Bayesian network classifiers. Mach. Learn. 29, 131–163 (1997)
Gámez, J.A., Rumí, R., Salmerón, A.: Unsupervised naïve Bayes for data clustering with mixtures of truncated exponentials. In: Proceedings of the 3rd European Workshop on Probabilistic Graphical Models (PGM’06), pp. 123–132 (2006)
Gilks, W.R., Richardson, S., Spiegelhalter, D.J.: Markov chain Monte Carlo in practice. Chapman and Hall, London (1996)
Jain, A., Murty, M., Flynn, P.: Data clustering: a review. ACM Comput. Surv. 31(3), 264–323 (1999)
Karciauskas, G.: Learning with hidden variables: a parameter reusing approach for tree-structured Bayesian networks. Ph.D. thesis, Department of Computer Science. Aalborg University (2005)
Langseth, H., Nielsen, T.D., Pérez-Bernabé, I., Salmerón, A.: Learning mixtures of truncated basis functions from data. Int. J. Approx. Reason. 55, 940–956 (2014)
Langseth, H., Nielsen, T.D., Rumí, R., Salmerón, A.: Inference in hybrid Bayesian networks. Reliabil. Eng. Syst. Saf. 94, 1499–1509 (2009)
Langseth, H., Nielsen, T.D., Rumí, R., Salmerón, A.: Parameter estimation and model selection in mixtures of truncated exponentials. Int. J. Approx. Reason. 51, 485–498 (2010)
Langseth, H., Nielsen, T.D., Rumí, R., Salmerón, A.: Mixtures of truncated basis functions. Int. J. Approx. Reason. 53(2), 212–227 (2012)
Lauritzen, S.L., Wermuth, N.: Graphical models for associations between variables, some of which are qualitative and some quantitative. Ann. Stat. 17, 31–57 (1989)
Lowd, D., Domingos, P.: Naive bayes models for probability estimation. In: ICML ’05: Proceedings of the 22nd International Conference on Machine Learning, pp. 529–536. ACM Press, New York (2005)
Minsky, M.: Steps towards artificial intelligence. Comput. Thoughts, 406–450 (1961)
Moral, S., Rumí, R., Salmerón, A.: Mixtures of truncated exponentials in hybrid Bayesian networks. ecsqaru’01. Lect. Notes Artif. Intell. 2143, 135–143 (2001)
Morales, M., Rodríguez, C., Salmerón, A.: Selective naïve bayes for regression using mixtures of truncated exponentials. Int. J. Uncertain. Fuzziness Knowl. Based Syst. 15, 697–716 (2007)
Newman, D., Hettich, S., Blake, C., Merz, C.: UCI repository of machine learning databases (1998). http://www.ics.uci.edu/~smlearn/MLRepository.html
Pearl, J.: Probabilistic reasoning in intelligent systems. Morgan-Kaufmann, San Mateo (1988)
Romero, V., Rumí, R., Salmerón, A.: Learning hybrid Bayesian networks using mixtures of truncated exponentials. Int. J. Approx. Reason. 42, 54–68 (2006)
Rumí, R., Salmerón, A.: Approximate probability propagation with mixtures of truncated exponentials. Int. J. Approx. Reason. 45, 191–210 (2007)
Sahami, M.: Learning limited dependence Bayesian classifiers. In: Second International Conference on Knowledge Discovery in Databases, pp. 335–338 (1996)
Schwarz, G.: Estimating the dimension of a model. Ann. Stat. 6, 461–464 (1978)
Shenoy, C., Shenoy, P.: Bayesian Network Models of Portfolio Risk and Return. In: Abu-Mostafa, W.L. Y.S., LeBaron, B., Weigend, A. (ed.) Computational Finance, pp. 85–104. MIT Press, Cambridge (1999)
Shenoy, P.P., West, J.C.: Inference in hybrid Bayesian networks using mixtures of polynomials. Int. J. Approx. Reason. 52(5), 641–657 (2011)
Smyth, P.: Model selection for probabilistic clustering using cross-validated likelihood. Stat. Comput. 10(1), 63–72 (2000)
Webb, G.I., Boughton, J.R., Wang, Z.: Not So Naive Bayes: Aggregating One-Dependence Estimators. Machine Learn. 58, 5–24 (2005)
Witten, I.H., Frank, E.: Data Mining: Practical Machine Learning Tools and Techniques, 2nd edn. Morgan Kaufmann, Burlington (2005)
Acknowledgments
This work has been supported by the Spanish Ministry of Science and Innovation through projects TIN2010-20900-C04-02,03, by the Regional Ministry of Economy, Innovation and Science (Junta de Andalucía) through projects P08-RNM-03945 and TIC2011-7821, and by ERDF funds.
Author information
Authors and Affiliations
Corresponding author
Additional information
A preliminary version of this paper was presented at the PGM’06 Workshop.
Appendix: Algorithms
Appendix: Algorithms






Rights and permissions
About this article
Cite this article
Fernández, A., Gámez, J.A., Rumí, R. et al. Data clustering using hidden variables in hybrid Bayesian networks. Prog Artif Intell 2, 141–152 (2014). https://doi.org/10.1007/s13748-014-0048-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13748-014-0048-3