Abstract
A water-soluble Cu (II) complex [(dien)Cu(μ-1,6-DAH)Cu(dien) (NO3)2](NO3)2 has been synthesized and its effect on the carrier model DNA structure and cancer cell line proliferation was investigated. In this regard, calf thymus DNA (CT-DNA) and human breast cancer cell line, T47D, were the targets. The effect of the complex on DNA structure was investigated by means of UV/vis, fluorescence and circular dichroism (CD) spectroscopic techniques as well as dynamic light scattering (DLS), zeta potential analysis and docking assay for more analysis. The UV–vis absorption spectra of complex with DNA showed a slight red shift and hypochromic effect, which indicated the intercalation and electrostatic effect of complex with CT-DNA. Using ethidium bromide (EB) as a probe in fluorescence studies revealed that complex can quench the EB–DNA fluorescence emission at different temperatures. Besides, the far UV–CD studies displayed that the complex induces changes in the secondary structure of CT-DNA and can increase the melting temperature of DNA up to 14 °C. The DLS and zeta potential measurements confirmed the electrostatic interaction of complex with the negatively charged DNA and subsequent DNA condensation. Besides, computational studies reflect that major and minor groove binding are two modes of interaction between complex and DNA. On the other hand, growth inhibition of the complex toward T47D cell line was measured using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay, which showed no cytotoxic properties.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
It has been reported that a wide range of agents including pigments [1], natural products [2], anionic lipids, polymers, dendrimers, polypeptides and nanoparticles are able to induce the condensation of DNA [3–6]. For example, cationic alkylammonium ligands displace Na+ cations, bind to DNA and condense the lipid–DNA complex [7]. The size of DNA aggregates can be affected appreciably by polymers rather than monomers [8] since it is found that the structures and dimensions of cationic polymers/DNA complexes are sensitively dependent on the condensing agents. In another study, it is reported that the DNA in the presence of 5-polyamidoamine (G5 PAMAM) cationic dendrimer particles is condensed to a much higher degree than in its absence. Moreover, high degree of cooperativity in dendrimer-induced DNA condensation and electrostatic complexation of the nanoparticle-embedded dendrimer leading to the assembly of gold nanoparticles on DNA has been reported [9, 11]. The interaction of the major protein component of the amyloid deposition in Alzheimer’s disease [39–43 residue peptide-amyloid β (Aβ)] with DNA indicates that Aβ induces double-stranded DNA condensation in vitro, and the condensation is time-dependent [10].
Some of transition-metal complexes such as cobalt (III) [10–13, 15], copper (II) [11] and ruthenium (II) [13, 14] Cu (II) and Ni (II) [16] Pt (II), Pd (II) [17] have displayed a remarkable capability for DNA binding and condensing. In addition, the impact of metal complexes on DNA condensation is more pronounced than that of purely organic condensing agents, in that [Co (NH3)6]3+ is four times more efficient than spermidine although they have the same charge (3+) [18]. Small cationic porphyrin molecules bind to DNA double helical structures and are stabilized by two major binding modes [19–22]: intercalating between stacked base pairs and groove binding by non-covalent interaction.
The two binding modes are both capable of altering the geometry of a nucleotide and subsequent structural changes of DNA. Some investigations showed that divalent metal complexation not only can lead to DNA condensation [23] or aggregation [24], cleavage [25], but also can alter its secondary or tertiary structure [26–28]. This makes the metal complex serve as a potential modulator of DNA function. In other studies metal complexes such as organo-tin (IV) and -Ni (II) were screened for antibacterial activities [29, 30].
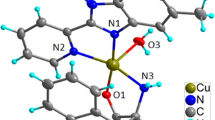
Copper-containing proteins are involved in various processes in living systems. Hemocyanin (O2 transport), tyrosinase (hydroxylation of monophenols and oxidation of catechols), and catechol oxidase (oxidation of catechols) are classified as type 3 copper proteins and have magnetically coupled binuclear copper (II) centers at their active sites [31–33]. In consideration of all of the stated factors, in this study we have designed a novel homobinuclear copper (II) complex of formula [(dien)Cu(μ-1,6-DAH)Cu(dien) (NO3)2](NO3)2, where μ-1,6-DAH = 1,6-diaminohexane as structural models for type III copper enzymes like tyrosinase. The rationale for this design is that copper has been known as an essential bioelement but its biological role(s) has been recognized only in the last decades due to the rapid development of bioinorganic chemistry and successful interaction between model complexes and protein biochemistry [31]. Also, bimetallic cores are becoming increasingly important to which may form a polynuclear complex ion with high positive charges which is beneficial for DNA condensation.
In this work, structural information regarding the interaction of a metal complex (as shown in Fig. 1) with CT-DNA were obtained by UV–vis absorption, circular dichroism and fluorescence spectroscopies as well as dynamic light scattering, zeta potential and molecular docking. In this way, binding constants, modes of interaction, thermodynamic parameters and morphology characteristic of DNA complex structure were determined. In the other experiment, cytotoxicity effect of complex on T47D cell line (as a model cancer cell line) was studied by MTT assay.
Materials and methods
Materials
Copper (II) nitrate trihydrate, diethylenetriamine, 1,6-diaminohexane and DOPA (3,4-dihydroxy phenyl alanine), and EDTA were purchased from Merck Company and used without further purification. Solvents were AR grade and were just distilled before use. Ethidium bromide, RPMI, MTT, trypsin–EDTA, streptomycin, and penicillin were obtained from Gibco (USA).
Methods
Preparation of [(dien)Cu(μ-1,6-DAH)Cu(dien) (NO3)2](NO3)2
A 0.54 ml (5 mmol) amount of diethylenetriamine in 15 ml distilled ethanol was added dropwise to a solution of 1.208 g (5 mmol) of copper (II) nitrate trihydrate in ethanol (20 ml) under constant stirring over a period of 2 h. The stirring continued for another 5 h and the dark blue precipitate formed was completely dissolved by adding 2.5 ml of water. The volume of reaction mixture was reduced to 16 ml at 40 °C and mixture was next cooled to the room temperature and dissolved 0.29 g (2.5 mmol) of 1,6-diaminohexane in 15 ml ethanol was added within an hour with constant stirring. The stirring further continued at 40 °C for another 4 h and the solution was stirred and concentrated to 18 ml at 40 °C. The precipitate formed, was then dissolved by adding 2 ml water and 8 ml acetonitrile. It was then stirred for a few hours and filtered. Dark blue needle-shaped crystals which are made after 5 days by diffusion of acetone into filtrate were washed with acetone and dried in an oven at 40 °C and used in X-ray investigations. The complex crystallized as anhydrate with yield of 1.4 g (80.3 %) and decomposes at 189–192 °C. The complex gives satisfactory elemental analysis for C14H42N12O12Cu2 (697.56): C, 24.10; H, 6.07; N, 24.10; Cu, 18.22 %. Found: C, 24.11; H, 6.08; N, 24.08; Cu, 18.20 %. Solid state FT-IR spectroscopy of the above complex (Fig. 1) shows characteristic stretching bands at 3,330 and 3,263 cm−1 (NH2 bonding), 1,384 and 1,315 cm−1 (monodentate NO −3 ), 1,763 cm−1 (uncoordinated NO3 −), 420 cm−1 (Cu–N) and 524 cm−1 (Cu–O). Molar conductance measurement in water for the complex is 667 Ω−1 mol−1 cm2 indicating 1:4 electrolytes. Electronic spectra exhibit the characteristic broad d–d band of weak intensity centered at 571 nm [34]. The absorption bands at 226 and 243 nm could be assigned to π → π* and n → π* transition of counterion nitrate [35].
DNA preparations
Purification of calf thymus DNA was done according to our previous article [36].
Spectroscopy studies
The UV–vis absorption spectra of the complex and complex–DNA were measured with a Cary spectrophotometer, 100 Bio-model, with jacketed cell holders.
Since the intrinsic fluorescence of DNA is very weak, therefore using extrinsic fluorescent probes is the complementary method for its detection. The fluorescence of EB can be enhanced by its intercalation between the base pairs of DNA [37]. Accordingly different concentrations of complex were added (0–300 μM) to the solution of EB and DNA (with the concentration of 2 and 60 μM, respectively), and fluorescence intensity changes were studied at 471 nm excitation wavelength (maximum quantum yield for ethidium bromide) at 37 °C by using a Hitachi spectrofluorimeter, MPF-4 model, equipped with a thermostatically controlled cuvette compartment. In this approach, emission intensities of complex as control were negligible at highest applied concentrations.
Circular dichroism spectrum is another useful method for monitoring the structural changes of DNA upon interaction with DNA in the region of 200–320 nm. UV–vis CD spectral changes were analyzed by an Aviv model 215 Spectropolarimeter (Lakewood, NJ, USA) from interaction of complex (0–32 μM) with 1.06 mM DNA at 37 °C. For this, water and water–complex spectra were subtracted from DNA and DNA–complex samples, respectively.
In temperature-scanning spectroscopy, Theta 240 profiles of DNA and DNA–nanoparticles which describe the thermal denaturation of DNA were monitored in the sample cells contained 2.12 mM DNA and 32 μM complex.
Besides, the hydrodynamic diameters of DNA were measured using dynamic light scattering (DLS). DLS was measured with an argon ion laser set at 488 nm and a fixed 90° scattering angle (Malvern Instrument, Worcestershire, United Kingdom). In this experiment, complex was added to DNA solution (2 mM) with the concentration range from 0 to 120 μM.
In another study, zeta potential was measured in deionized water solution by adding DNA solution (2 mM) to complex (0–50 μM). After incubation of each mixture at 37 °C for 60 min, the mean of triplicate experiments was taken as the final result and reported.
Docking study
Docking refers to a computational scheme that tries to find the binding energy and binding site correspond to interaction between ligand and macromolecule. The PDB file of B-DNA as double strand (1ZFO) was extracted from PDB data bank (http://www.rcsb.org), and free energy of docking compounds was calculated. Moreover, ligand was drawn by Chemoffice 2007 and molecular structure was optimized using HyperChem 7.0. Polar hydrogens were added, and Gasteiger charges were computed for the porphyrin as ligand. For other derivatives, all water molecules were removed and Kollman charges and solvation parameters were added. Based on the atom types, the appropriate maps need to be calculated for the macromolecule. Grid maps of 100 × 70 × 60 points with a grid-point spacing of 0.375 Å were generated using the Auto Grid tool of ADT. The 100 genetic algorithm (GA) runs were performed with the following parameters: population size of 150, maximum number of 2.5 × 105 energy evaluations, maximum number of 27,000 generations, an elitism of 1, a mutation rate of 0.02 and a crossover rate of 0.8 were selected. The resulting conformations were clustered using a root-mean-square deviation (rmsd) of 0.5 Å.
Cytotoxic measurements
Cell culture
In RPMI medium, the human breast cancer cell lines T47D were grown. This medium was supplemented with l-glutamine (2 mM), streptomycin, and penicillin (5 μg/ml), 10 % heat-inactivated fetal calf serum. Cells were maintained in 5 % CO2 and 95 % air atmosphere incubator at 37 °C.
MTT assay
Because of probable inhibition growth of human breast cancer cell line, T47D by complex, MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide], could be used to test cytotoxicity of complex and cell viability. In the mitochondria the conversion of the soluble yellowish MTT to the insoluble purple formazan by active mitochondrial of living cells is an assay for measurement of cell proliferation.
The cells were harvested with trypsin–EDTA when 90 % confluence was reached in T25 flasks, washed with PBS buffer subsequently 1 × 104 cells were plated into each well of 96-well plates in 180 μl of growth medium and left to attach to the plates. Then, harvested cells were seeded with different concentrations of the sterilized complex (0–0.3 mM) and incubated for 24 h. Two hours to the end of incubations, 25 μl of MTT working solution (5 mg/ml in PBS) was added into wells being assayed. At the end of the incubation period the converted dye is solubilized with a solution containing 10 % SDS and 50 % DMF (for 2 h at 37 °C under dark conditions) and absorbance of the converted dye was measured at a wavelength of 570 nm with background subtraction with a multiwell scanning spectrophotometer (ELISA reader, Model Expert 96, Asys Hitchech, Austria). Percent survival was calculated as the percent of OD value of the study groups relative to that of the untreated control wells. Each experiment was repeated three times and for each concentration performed in triplicate format. For each concentration mean values ± SE was determined.
Results and discussion
UV–vis studies
UV–vis spectroscopy is an effective method to investigate the binding mode of DNA with metal complexes [38]. Thus, in order to provide information for the possibility of binding of complex to CT-DNA, the absorption spectra of complex (30 μM) was measured in the range of 190–290 nm about 2 min after addition of DNA solution (60 μM). UV–vis spectra were recorded in the absence and presence of different concentrations of DNA and shown in Fig. 2. The absorption maximum of DNA–complex system (curve c) was located at approximately 204 nm, which is comparable with that calculated from the sum of absorbance of the free complex and the DNA. The results demonstrated that the measured spectra were less intense than the stimulated one (curve d). Furthermore, there was a slight red shift at the maximum wavelength of the measured spectra. This hypochromic effect on the spectra of the DNA–complex mixture, as well as the noted slight red spectral shift, suggested that the interaction of the complex with CT-DNA takes place by stacking and electrostatic effect [39].
Fluorescence studies
Competitive intercalation between EB and positively charged complexes can result in quenching of the fluorescence [39]. Therefore, displacement of EB could use as an assay for the interaction between DNA and complex. As shown in Fig. 3, upon addition of complex to a DNA–EB solution, the fluorescence intensity decreased dramatically because of replacement of a great mass of EB by the complex from a hydrophobic environment to water solution [39].
Binding property of DNA complexes
Figure 3a–c shows a drastic quenching of DNA–EB solution fluorescence at different concentrations of complex at 20, 37, and 42 °C, respectively. The fluorescence quenching data were analyzed by the Stern–Volmer [40, 41]:
where F 0 and F are the steady-state fluorescence intensities in the absence and presence of complex, respectively, K sv is the Stern–Volmer quenching constant and [Q] is the concentration of complex.
By using Fig. 4, the values of K sv at different temperatures were calculated and shown in Table 1. According to the classical Stern–Volmer equation, the quenching plot illustrate that the quenching of EB bound to DNA by the complex is in good agreement with the linear Stern–Volmer equation, which also proves that the complex binds to DNA [41]. As shown the quenching constant K sv values enhance with increasing of temperature which indicated that the probable quenching mechanism of DNA–EB solution is a dynamic quenching procedure and complex between complex and DNA–EB may be formed.
Binding parameters
The binding constant (K) and the number of binding sites (n) between complex with DNA can be calculated using the Eq. 2 for the quenching experiment [40, 41].
If we plot the log [(F 0 − F)/F] versus log [Q], it gives a straight line, in which slope equals to n (the number of binding sites) and the intercept presents log K. The results revealed multiple class of binding site on DNA for complex exist in which it reacts with DNA through electrostatic interaction and intercalation.
The values of K and n at 20, 37, and 42 °C are listed in Table 1. The binding constant of complex increases with increasing temperature, which indicates that the stability of the DNA–complex complexes is strengthening with increasing the temperature.
Analysis of interaction force between DNA and complex
The signs and magnitudes of thermodynamic parameters for DNA reactions account for the main forces contributing to the DNA stability. If we suppose that the enthalpy change (ΔH°) over the above temperature range is minimum, then the thermodynamic parameters such as ΔH° and ΔS° could be determined using the van’t Hoff Eq. 3:
where K is the binding constant at the corresponding temperature and R is the gas constant. The slope and Y-intercept of the plot of ln K versus 1/T determine the ΔH° and ΔS°, respectively (Fig. 5). Then the Gibbs free energy (ΔG°) is estimated from the following Eq. 4:
The calculated thermodynamic parameters are summarized in Table 1. The binding parameters between complex and DNA indicated that by increasing the temperature (from 20 to 42 °C), the affinity of binding of complex to DNA was increased, which is consistent with endothermic process of binding. The negative sign for ΔG° indicates spontaneous binding of complex to DNA. The positive value of ΔH° and ΔS° demonstrate that the binding of complex to DNA is an entropy-driven and endothermic process, which are associated with the electrostatic and stacking effect, respectively. It could also be explained that, owing to the binding of complex to DNA, some conformational changes are induced results to enhancing the stacking of the base pairs.
The presence of multivalent complex cations results in charge neutralization on the DNA backbone, which is accompanied by the collapse of linear DNA to a compact configuration [28]. The results prove that there is a strong interaction between complex and DNA which exerts a remarkable impact on DNA configuration.
CD studies
CD is a useful technique for monitoring conformational changes in DNA, which can occur as a result of changes in experimental parameters such as temperature and ligand binding. Therefore, further experiment was done by CD spectroscopy to investigate the changes in DNA conformation induced by complex in different concentrations. As shown in Fig. 6, the CD spectrum of CT-DNA shows a positive band at 275 nm owing to base stacking and a negative band at 245 nm owing to the helicity, which are characteristic of B-DNA [42].
Simple groove binding and electrostatic interaction of small molecules demonstrate less or no perturbation on the base stacking and helicity bands, while intercalation changes the intensities of both of the bands [43]. The complex solution has no CD spectrum when is free in the solution. When the complex was incubated with DNA, the CD spectra of DNA displayed a decrease of both positive and negative bands (Fig. 6). As seen in Fig. 6 after adding different concentrations of complex, the intensities of positive and negative bands decrease significantly, suggesting a strong indicator of classical intercalation. It can be concluded that the presence of complex may significantly change the helicity of DNA. These result in alterations in the orientations of DNA bases, tilted them, changes of DNA base stacking and finally role as an intercalator. These data are broadly consistent with decreasing of EB fluorescence intensity. For confirmation of stacking effect of complex, melting temperature of DNA in the absence and presence of complex was investigated.
DNA thermal denaturation studies
Thermal behaviors of DNA in the presence of complex can provide useful information into DNA conformational changes and interaction strength of complex with DNA. It is already known that, when the temperature increases for the DNA solution, the double-stranded DNA gradually dissociates to single strands resulting therefore in a hyperchromic effect on the absorption spectra of DNA. In order to identify the effect of complex on the structure of DNA, the melting temperature (T m), (the temperature where half of the total base pairs are unbounded), is usually measured. It has been reported that the intercalation of metallo-intercalators generally lead to a drastic enhancement of T m [44, 45]. The melting curves of DNA in the absence and presence of the complex are presented in Fig. 7. It is shown that DNA in the absence of the complex has a T m value of 73.9 ± 1 °C, whereas the observed melting temperature of DNA in the presence of complex successively increased to a value of 88 ± 1 (°C). The increasing of T m for DNA in the presence of complex are comparable to those observed for classical intercalators [36].
Dynamic light scattering
DLS measurements were used to investigate the size distribution of the DNA particles in solution induced by complex. Figure 8 shows the distribution of hydrodynamic radius of the DNA particles measured at a scattering angle of 90° at 25 °C. The apparent mean diameter of the condensed DNA particles was about 1,000 nm, which confirms existence of condensed DNA particles in solution.
Zeta potential measurements
Zeta (z) potential serves as an indication of the surface charge of macromolecular, which can be considerably affected by aggregation and distribution of the particles [46]. The zeta potentials changes of DNA particles at different concentrations of complex are listed in Table 2. It can be seen that as the concentration of complex was increased, the negative surface charges of the DNA particles decreased; therefore, the zeta potential increased with increasing concentration of complex from −29.81 to −3.44 mV. This change in zeta potential value confirms that the condensation is occurring when complex was added to DNA solutions. Surface negative charge distribution is a crucial factor for stabilizing the DNA particles by electrostatic repulsion and therefore prevents them from condensing. Increasing positive charge causes electrostatic interaction with negatively charged DNA and leads to the DNA condensation [47, 48].
Docking results
Figure 9a, b show a fraction of B-DNA as double strand and the most negative ranks of the ligand that shows the position of ligand bound to DNA. List of docking first 13 ranks results that are the cluster number, docking energy and number of run in one position are presented in Table 3. As we see from Table 3, there are different sites and different docking energies. Last column of table shows number of runs that happen in one position which demonstrates probability of locating of ligand on that site. For example cluster 1 has 2 runs in only one position while cluster 4 and 11 have the 34 and 71 number of runs in a cluster, respectively. It seems that the possibility of cluster 9 is more than 2 and it is higher than 1 which indicates that two major and minor groove binding sites may be preferred. Minor groove has more negative energy (more favorable) than major groove. So the first docking site can be related to first binding site of fluorescence results. In docking study, there is no staking interaction between ligand and bases. It may due to the larger size of ligand which hinders diffusion between base layers.
Cytotoxicity measurements of the complex
The in vitro cytotoxicity property of the synthesized complex was studied by testing them on human breast cancer cell line, T47D. As presented in Fig. 10, the complex did not exhibit obvious cytotoxicity towards the T47D cell line at different concentrations. In this study, various concentrations of complex ranging from 0 to 0.3 mM were used to culture the tumor cell lines for 24 h (Fig. 10). Even when the concentration of complex was increased to 0.3 mM, the viabilities of each of the cell lines were still above 94 %. The results indicate that the cytotoxicity of complex is suitably low. Therefore, the toxicity of this complex to human breast cancer cell line of T47D is negligible and it might be concluded that this cancer cell line is resistance to the designed copper complex.
Conclusion
In this work, we explored the binding interaction of a new copper complex with DNA in deionized water by different spectroscopic techniques. The binding constants and number of binding sites of complex with DNA were measured at different temperatures, and the thermodynamic parameters were calculated as well in order to found possible interaction between complex–DNA and better sight in rational design gene vector complexes. It was found that intercalation and electrostatic effect play a major role in the binding of complex to DNA. The intercalative binding of complex with DNA was explored by our data from UV–vis absorption spectra, fluorescence spectra, circular dichroism spectra and melting temperature determinations. The electrostatic mode of interaction between DNA and complex was deduced from DLS and zeta potential experiments. For more investigation docking was performed and these modes of interaction were approved.
The biological significance of this work may be evident since DNA contains the genetic instructions used in the development and functioning of all known living organisms. Therefore, the interaction of the complex can provide useful information to better design gene vector complexes with fewer side effects.
References
S. Nafisi, A.A. Saboury, N. Keramat, J. Neault, H.A. Tajmir-Riahi, J. Mol. Struct. 827, 35 (2007)
C.D. Kanakis, P.A. Tarantilis, M.G. Polissiou, S. Diamantoglou, H.A. Tajmir-Riahi, J. Biomol. Struct. Dyn. 22, 719 (2005)
N. Korolev, N.V. Berezhnoy, K.D. Eom, J.P. Tam, L. Nordenskiçld, Nucleic Acids Res. 37, 7137 (2009)
J.P. Behr, Acc. Chem. Res. 26, 274 (1993)
V. Vijayanathan, T. Thomas, T.J. Thomas, Biochem. 41, 14085 (2002)
E.K. Esbjçrner, P. Lincoln, B. Nord, Biochem. 47, 1732 (2008)
D. Matulis, V.A. Bloomfield, J. Am. Chem. Soc. 124, 7331 (2002)
L. Liu, YL. Yang, C. Wang, Y. Yao, YZ. Ma, S. Hou, XZ. Feng, Colloids Surf. B Biointerf. 75 (2009) 230
K. Fant, E.K. Esbjörner, P. Lincoln, B. Nordén, Biochemistry 47, 1732 (2008)
Y. Haijia, R. Jinsong, Q. Xiaogang, J. Biophys. 92, 185 (2007)
Y.C. Liu, H.L. Chen, H.K. Lin, W.L. Liu, Y.W. Chou, S.C. Lo, C.-H. Tai, Biomacromolecules 6, 3481 (2005)
X.G. Meng, L. Liu, C.S. Zhou, L. Wang, C.L. Liu, Inorg. Chem. 47, 6572 (2008)
E. Musatkina, H. Amouri, M. Lamoureux, T. Chepurnykh, C. Cordier, J. Inorg. Biochem. 101, 1086 (2007)
B. Sun, J.X. Guan, L. Xu, B.L. Yu, L. Jiang, J.F. Kou, L. Wang, X.D. Ding, H. Chao, L.N. Ji, Inorg. Chem. 48, 4637 (2009)
P. Nagababua, J. Naveena lavanya lathaa, M. Rajeshb, S. Satyanarayanaa J. Iran. Chem. Soc. 6, 145 (2009)
N. Ramana, L. Mitub, A. Sakthivela, M.S.S. Pandia, J. Iran. Chem. Soc. 6, 738 (2009)
M. Islami-Moghaddama, H. Mansouri-Torshizia, A. Divsalar, A.A. Saboury, J. Iran. Chem. 6, 552 (2009)
J. Pelta, F. Livolant, J.L. Sikorav, J. Biol. Chem. 271, 5656 (1996)
M.A. Sari, J.P. Battioni, D. Dupre, D. Mansuy, J.B. Lepecq, Biochem. Biophys. Res. Commun. 141, 643 (1996)
B.P. Hudson, J. Sou, J.D. Berger, D.R. McMillin, J. Am. Chem. Soc. 114, 8997 (1992)
M.A. Sari, J.P. Battioni, D. Dupre, D. Mansuy, J.B. Lepecq, Biochemistry 29, 4205 (1990)
K. Butje, K. Nakamoto, J. Inorg. Biochem. 39, 75 (1990)
C. Ma, V.A. Bloomfield, J. Biophys. 67, 1678 (1994)
J.G. Duguid, V.A. Bloomfield, J. Biophys. 69, 2642 (1995)
N. Raman, R. Jeyamurugan, B. Rajkapoor, L. Mitu, J. Iran. Chem. Soc. 7, 917 (2010)
V.V. Andrushchenko, S.V. Kornilova, L.E. Kapinos, E.V. Hackl, V.L. Galkin, D.N. Grigoriev, J. Mol. Struct. 225, 408 (1997)
E.V. Hackl, S.V. Kornilova, L.E. Kapinos, V.V. Andrushchenko, V.L. Galkin, D.N. Grigoriev, J. Mol. Struct. 229, 408 (1997)
H. Arkawa, R. Ahamad, M. Naoui, H.A. Tajmir-Rjahi, J. Biol. Chem. 275, 1050 (2000)
S. Shah, S. Ali, J. Shahzadi, Iran. Chem. Soc. 7, 59 (2010)
A.T. Çolak, F. Çolak, O.Z. Yeşilel, E. Şahin, J. Iran. Chem. Soc. 7, 384 (2010)
K. Selmeczi, M. Réglier, M. Giorgi, G. Speier, Chem. Rev. 245, 191 (2003)
D. Boghaei, A. Behzad, A. Bezaatpour, J. Mol. Catal. A-Chem. 241, 1 (2005)
C. Belle, K. Selmeczi, S. Torelli, J.L.C.R. Pierre, Chimie. 10, 271 (2007)
C.A. Bolos, G.S. Nikolov, L. Ekateriniadou, A. Kortsaris, D.A. Kyriakidis, Met.-Based Drugs 5, 323 (1998)
A.T. Chaviara, P.J. Cox, K.H. Repana, R.M. Papi, K.T. Papazisis, D. Zambouli, A.H. Kortsaris, D.A. Kyriakidis, C.A. Bolos, J. Inorg. Biochem. 98, 1271 (2004)
M. Rahban, A. Divsalar, A.A. Saboury, A. Golestani, J. Phys. Chem. 114, 5798 (2010)
A.G. Krishna, D.V. Kumar, B.N. Khan, S.K. Rawal, K.N. Ganesh, Biochim. Biophys. Acta 104, 1381 (1998)
G.S. Son, J.A. Yeo, M.S. Kim, S.K. Kim, A. Holmen, B. Akerman, B. Norden, J. Am. Chem. Soc. 120, 6451 (1998)
Y.N. Ni, D.Q. Lin, S. Kokot, Anal. Biochem. 352, 231 (2006)
S. Bi, C. Qiao, D. Song, Y. Tian, D. Gao, Y. Sun, H. Zhang, Sensor. Actuat. B 119, 199 (2006)
M. Chakraborty, A. Sengupta, D. Bhattacharya, S. Banerjee, A. Chakrabarti, Biochim. Biophys. Acta. 1804, 2016 (2010)
G. Psomas, A. Tarushi, E.K. Efthimiadou, Polyhedron 27, 27133 (2008)
J.W. Chen, X.Y. Wang, Y.G. Zhu, J. Lin, X.L. Yang, Y.Z. Li, Y. Lu, Z.J. Guo, Inorg. Chem. 44, 3422 (2005)
S.S. Jain, M. Polak, N.V. Hud, Nucleic Acids Res. 31, 4608 (2003)
J.L. Mergny, G. Duval-Valentin, C.H. Nguyen, L. Perrouault, B. Faucon, M. Rougee, T. Montenay-Garestier, E. Bisagni, C. Helene, C. Sci. 256, 1681 (1992)
R.M. Xing, X.Y. Wang, C.L. Zhang, Y.M. Zhang, Q. Wang, Z. Yang, Z.J. Guo, J. Inorg. Biochem. 103, 1039 (2009)
A. Olad, M. Khatamian, B. Naseri, J. Iran. Chem. Soc. 8, 141 (2011)
M. Hosseini-Sarvari, J. Iran. Chem. Soc. 8, 119 (2011)
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Derakhshankhah, H., Saboury, A.A., Bazl, R. et al. Synthesis, cytotoxicity and spectroscopy studies of a new copper (II) complex: calf thymus DNA and T47D as targets. J IRAN CHEM SOC 9, 737–746 (2012). https://doi.org/10.1007/s13738-012-0086-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13738-012-0086-3