Abstract
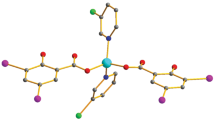
The title compound, diaqua(bissalicylato-κO)(bisnicotinamide-κN)zinc(II)][bis(triaqua (monosalicylato-κO)(mononicotinamide-κN)zinc(II)salicylate, includes three Zn(II) ions, four nicotinamide ligands, six salicylate ligands and eight coordinated aqua ligands in the asymmetric unit in complex structure. The geometry around one of the Zn(II) ions is a slightly distorted octahedron, of which the equatorial plane is formed by two carboxylate oxygens and two aqua oxygens, while the axial positions are occupied by two pyridyl nitrogen atoms. The other Zn(II) ions adopt fivefold coordinations with one carboxylate oxygen atom from salicylate ligand, one N atom from nicotinamide ligand and three oxygen atoms from aqua ligands. In addition, there are two salicylate anions in the unit cell that are not coordinated. They provide charge balance as counter-ions in the complex framework.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Metal complexes of aromatic carboxylic acids have been extensively studied for a few decades due to their synergetic or antagonistic effects on biological activity. Anti-inflammatory and antibacterial activity of metal carboxylates was found higher than that of the parent carboxylic acids [1]. In this study, we attempted to prepare a mixed ligand complex of Zn(II) with salicylic acid and nicotinamide. The motivation behind the choice of the ligands (Scheme 1) and the metal lies in their well-known biological importance. Benzoic acid and derivatives are widely used as antimicrobial agents in food and in dermatology, as a fungicidal treatment for fungal skin diseases in combination with salicylic acid [2–6]. Nicotinamide is a form of vitamin B3 and is an important component of several enzymes. Besides the usual complexes with metal ions, a number of mixed ligand nicotinamide complexes have been reported. Nicotinamide complexation appears to be a useful approach to increase the solubility of physiologically useful systems [7]. Zinc is an essential and beneficial element in human growth. This element is known to regulate the activity of over 300 metalloenzymes and it is a component of special proteins, called ‘‘zinc fingers’’ which participate in the reliable transfer of genetic information [8–10]. Zinc and its compounds have antibacterial and anti-viral activity and the wound-healing effect of zinc-containing ointments has been known for several centuries [11–13]. Zinc may be used as a therapeutic agent, it may act as an anti-sickling agent and play a role in the prevention of pain crisis in sickle-cell disease. Zinc was successfully used in the treatment of acrodermatitis enteropathica, Wilson’s disease, gastrointestinal disorders, infertility and other diseases [14–16].
There are many known organic ligand complex structures of Zn(II) ion, with coordination numbers of five or six, generally mononuclear and of mixed ligand type. There are few dinuclear or polynuclear complex structures of Zn(II), showing both five and six coordinations in the same framework structure where the metal ions are bonded to each other through a metal–metal bond or bridging a ligand molecule. In the several Zn(II)-carboxylate compounds, the bonding occurs through the acidic –OH group of carboxylic acid as monodentate [7, 17–22]. According to the X-ray structures available in literature, phenolic –OH group of aryl carboxylic acid does not participate in coordination and remains free. In some complexes, coordination is different as the carbonyl oxygen atom and acidic –OH group of carboxylic acid form a chelate structure [21]. The nicotinamide ligand generally participates in coordination through the hetero nitrogen atom of pyridine ring.
Here, we report the synthesis, spectroscopic, thermal and crystallographic properties of the mixed ligand complex {[Zn(na)2(sal)2(H2O)2][Zn(na)(sal)(H2O)3]2·2(sal)}. Considering that the carboxylate group has a versatile coordination behaviour [23] and Zn(II) has a flexible coordination sphere [24], the resulting complex may display an interesting supramolecular framework structure in addition to unprecedented biological properties.
Experimental
Materials and instrumentation
All chemicals used were analytical reagent products. ZnSO4·6H2O, salicylic acid and nicotinamide were obtained from Merck (Darmstadt, Germany). Elemental analyses (C, H, N) were carried out by standard methods. IR spectra were recorded in the 4,000–400 cm−1 region with a Perkin Elmer spectrum One FT-IR spectrophotometer using KBr pellets. Thermal analyses (TGA/DrTG, DTA) were performed by the Shimadzu DTG-60H system, in dynamic nitrogen atmosphere (100 mL/min) at a heating rate of 10 °C/min, in platinum crucibles as sample vessel, using α-Al2O3 as reference.
Preparation of the complex
In the first step, sodium salicylate was prepared in aqueous solution according to the following equation:
In the second step, zinc salicylate was prepared by adding a Zn(II) salt to the first solution:
Finally, the solution of nicotinamide (2 mmol) in distilled water (75 mL) was added dropwise to a stirred solution of Zn(C7H5O3)2(H2O) n (1 mmol) in hot distilled water (75 mL):
The resulting solution was allowed 25–27 days for crystallization at room temperature. The crystals formed were filtered and washed with cold water and acetone and dried in vacuo. The yield of complex is 65%.
Results and discussion
According to the analytical results, the complex contains six moles of salicylate, four moles of nicotinamide and eight moles of aqua ligands per formula unit.
X-ray crystallographic data revealed that, two of the three ZnII ions are penta-coordinated, while the third is hexa-coordinated. The fivefold coordinations are completed with one salicylate, one nicotinamide and three aqua ligands, while the sixfold coordination is completed with two salicylate, two nicotinamide and two aqua ligands apiece. All of the ligands are bonded monodentately [21–25] and two salicylate ions remain as counter-ion in the structure.
FT-IR spectra
The absorption bands observed in the range of 3,450–3,050 cm−1 correspond to the asymmetric and symmetric stretching vibrations of water molecules. The peaks for the N–H stretches of primary amides are strong in the range of 3,340–3,250 cm−1. The N–H bending vibrations appear approximately in the range of 1,520 cm−1. Normally, the benzoate complexes give rise to strong bands responsible from the C=O stretching for which conjugation between the carbonyl group and the amide nitrogen causes small frequency shifts [26]. The strong bands observed around 1,645 cm−1 are assigned to this mode. This band remained almost in the same wavenumber range as the amide group of the free na ligand, indicating that the na ligand does not coordinate through the amide group. Pyridine ring vibration of free nicotinamide at 1,535 cm−1 shifts to 1,450 cm−1 in the complexes indicating that the pyridine ring is coordinated. The main difference in the spectrum of salicylic acid is that the C=O stretching vibration of the carboxyl group at 1,720 cm−1 shifts to lower frequency in the metal complex. The carboxylate peaks in the metal complex appear in the range of 1,597 cm−1. This shows that the coordination takes place through the carboxyl group [27].
The –OH bending peak of salicylic acid in the metal complex is observed around 1,265 cm−1. The low intensity bands in the region of 600–400 cm−1 are attributed to M–N and M–O vibrations [27, 28].
The frequency of the asymmetric carboxylate vibration, ν as(COO−), and the magnitude of the separation between the carboxylate stretches, Δ = ν as(COO−)−ν s(COO−) are often used as spectroscopic criteria to determine the mode of the carboxylate binding. Generally, the following order is proposed for divalent metal carboxylates:
In the monodentate coordination, the redistribution of the electron density takes place and the shift of ν as(COO−) to higher wavenumbers is observed in comparison with the ionic group, increasing the value of Δ. On the contrary, the chelating coordination shifts the position of the asymmetric carboxylate stretch to lower wavenumbers in comparison with the ionic group and lowers the value of Δ. In general, the comparison of the Δ value of the respective complex with the Δ value of the particular sodium salt should be used for the assignment of the following guidelines: (i) bidentate chelating coordination occurs when Δ(COO−)complex > Δ(COO−)sodium salt, (ii) monodentate coordination is characterised by Δ(COO−)complex < Δ(COO−)sodium salt [29].
The ν s(COO−) peak is located at 1,597 cm−1, while the ν as(COO−) peak is observed at 1,393 cm−1 for the studied complex. The shift (Δ) between the ν as and ν s bands of (COO−) group is 204 cm−1 for complex, which is more than that for the sodium salicylate salt (159 cm−1) indicating that monodentate carboxylate group exists [22, 30].
Thermal analysis
The TG/DrTG and DTA curves for the homo-trinuclear title compound are given in Fig. 1. This complex is stable in air up to 159 °C. Further heating causes the release of 8 moles of coordinated water molecules by an accompanying endothermic effect in the DTA curve at 176 °C (mass loss: exp. 8.01%; calcd. 8.71%). The anhydrous complex is stable in the range of 182–233 °C and decomposes with melting starting at 235 °C and finishing at 330 °C, with the corresponding DTA peak at 266 °C. The four moles of neutral nicotinamide ligands are destroyed in this step (mass loss: exp. 26.01%; calcd. 29.50%). The next three decomposition steps belong to removing of anionic salicylate ligands in the range of 345–720 °C with DrTG peaks at 385, 419, 490 °C. Finally, the decomposition product, ZnO, was obtained as identified by IR spectroscopy comparing with the corresponding spectrum of the pure oxide (mass loss: exp. 13.88%; calcd. 14.90%), likewise reported elsewhere [17–20, 30].
Crystallography
Diffraction data were collected at 296 K using a STOE IPDS-II diffractometer; in all these cases graphite-monochromated MoKα radiation (λ = 0.71073 Å) was employed. Other details of cell data, data collection and refinement are summarized in Table 1.
The structure was solved by direct methods using SHELXS-97 and refined by full-matrix least-squares methods on F2 using SHELXL-97 [31] from within the WINGX [32] suite of software. All non-hydrogen atoms were refined with anisotropic parameters. Water H atoms were located in difference maps and refined subject to a DFIX restraint of O–H = 0.83(2) Å, and with U iso(H) = 1.5U eq(O). All other H atoms were located from difference maps and then treated as riding atoms with C–H distances of 0.93–0.97 Å, O–H distances of 0.82 Å and N–H distances of 0.86 Å, and with U iso(H) = 1.2Ueq(C, N, O). Supramolecular analyses were made and the diagrams were prepared with the aid of PLATON [33, 34]. Details of hydrogen-bond dimensions are given in Table 2.
Diaqua(bissalicylato-κO)(bisnicotinamide-κN)zinc(II)][bis(triaqua(monosalicylato-κO) (mononicotinamide-κN)zinc(II)salicylate
The complex crystallizes with Z = 1/2 in the space group P1 and is two-dimensional. In the supramolecular structure given in Fig. 2, it is possible to identify four independent one-dimensional sub-structures: One is built from an N–H···O hydrogen bond, augmented by two O–H···O hydrogen bonds, where all three interactions utilize a single acceptor (Table 2). The other one is built from just a single N–H···O hydrogen bond (Table 2). The third sub-structures are built from two N–H···O hydrogen bonds (Table 2); and finally, there is a fourth one-dimensional sub-structure built from six O–H···O hydrogen bonds (Table 2) and two π···π stacking interactions.
Water oxygens [O4 in the molecule at (x, y, z) and O2 in the molecule at (1 + x, y, z)] both act as hydrogen bond donors to the O14 atom in the molecule at (1 − x, −y, 1 − z), while O2 at (x, y, z) and O4 at (−1 + x, y, z) in turn act as donors to O14 at (−x, −y, 1 − z). Similarly, water oxygen O3 in the molecule at (x, y, z) acts as a hydrogen-bond donor, via H3A, to atom O13 in the molecule at (1 − x, −y, 1 − z) (Fig. 3).
Atom O8 at (x, y, z) acts as hydrogen-bond donors, respectively, to atoms O11 at (1 + x, y, z) and O12 at (1 − x, −y, 2 − z). These two hydrogen bonds are individually generated by translation of two independent anti-parallel C(8) chains, one built exclusively of type 1 molecules and the other containing only type 2 molecules. Combination of the two O–H···O hydrogen bonds generates a chain of edge-fused rings. Atom N4 in the reference molecule at (x, y, z) acts as a hydrogen-bond donor to atom O10 in the molecule at (x, −1 + y, z), so forming a chain of rings running parallel to the b axis (Fig. 4).
The pyridine ring N1/C8/C9/C10/C11/C12 in the molecule at (x, y, z) makes dihedral angles of only 7.9(2)º with the phenyl ring C1–C6 in each of the two molecules at (−x, 1 − y, 1 − z) and (1 − x, 1 − y, 1 − z). The ring-centroid separations are 3.969 (4) and 3.852 (4) Ǻ, respectively, and the interplanar spacings are ca 3.55 and 3.32 Ǻ, corresponding to ring offsets of ca 2.01 and 1.61 Ǻ (Fig. 5).
The combination of the chains suffices to generate a two-dimensional structure of considerable complexity.
Conclusions
There are three metal nucleus in unit cell of the complex. While, just one Zn(II) cation nucleus has sixfold, other nucleus have fivefold coordination. The sixfold Zn(II) structure contains two moles of salicylate and nicotinamide and aqua ligands, while fivefold Zn(II) structures include one mole of salicylate and nicotinamide and three moles of aqua ligands, concurrently there are two moles of salicylate anions as counter-ion to provide charge-balance in the neutral complex framework. The fivefold ZnII layer and sixfold ZnII layer in complex framework were showed in Fig. 6.
The following conclusions can be drawn from the results of thermal decomposition, IR and X-ray spectroscopic studies on the title complex:
-
1.
The thermal decomposition of the title complex takes place in three steps: dehydration, elimination of the nicotinamide ligand and decomposition of the salicylato ligand. This decomposition manner is similar to those previously reported for some mixed nicotinamide–salicylate metal complexes [27, 28, 30]. Early releasing of the nicotinamide before the salicylate ligand by volatilisation may be due to non-ionic bonding of this ligand with Zn(II) ion.
-
2.
IR spectroscopy helped determination of carboxylate binding mode. In the complex, all ligands coordinate to Zn(II) monodendately.
-
3.
The unit cell contains three Zn(II) ions. The geometry around one of them is a slightly distorted octahedron, while the other two Zn(II) ions are penta-coordinated. Hexa-coordinated Zn(II) ions and penta-coordinated Zn(II) ions constitute layers along the c axis and ab plane, respectively. The layers are non-interpenetrating but perpendicular to each other (Fig. 7).
The layers are bonded to each other by second interactions. The layers settled in opposite directions to each other. The coordination gaps occur between the layers.
The supramolecular structure shown by the hexa- and penta-coordinated Zn(II) ions here is unusual and may suggest the potential use of this material to host for small guests.
Supplementary material
CCDC 759975 contains the supplementary crystallographic data for this paper. These data can be obtained free of charge from The Cambridge Crystallographic Data Centre via http://www.ccdc.cam.ac.uk/data_request/cif.
References
J.R.J. Sorenson, V. Kishore, A. Pezeshk, L.W. Oberley, S.W.C. Leuthauser, T.D. Oberley, Inorg. Chim. Acta 91, 285 (1984)
G. Cavigiolio, L. Benedetto, E. Boccaleri, D. Colangelo, I. Viano, D. Osella, Inorg. Chim. Acta 305(1), 61 (2000)
U. Bruehlmann, E. Hayon, J. Am. Chem. Soc. 96(19), 6169 (1974)
J.R.J. Sorensen, in Metal Ions in Biological Systems, vol. 14, ed. by H. Sigel (Marcel Dekker), New York, 1982, p.77
C.A. Rice-Evans, N.J. Miller, G. Paganga, Free Radical Biol. Med. 20, 933 (1996)
A. Hossaini, J.J. Larsen, J.C. Larsen, Food Chem. Toxicol. 38, 319 (2000)
D.A. Köse, B. Zümreoglu-Karan, C. Unaleroğlu, O. Şahin, O. Büyükgüngör, J. Coord. Chem. 18, 2125 (2006)
B.L. Vallee, D.S. Auld, Biochemistry 24, 5647 (1990)
E. Kimura, T. Koike, Adv. Inorg. Chem. 44, 229 (1997)
W. Kaim, B. Schwederski, Bioinorganic Chemistry: Inorganic Elements in the Chemistry of Life (Wiley, Chichester, 1994)
E.J. van Hoogdalem, J. Eur. Acad. Dermatol. Venereol 11, 13 (1998)
E. De Clercq, Metal-based drugs 4, 173 (1997)
O. Yamamoto, Int. J. Inorg. Mater. 3, 643 (2001)
Y.H. Cho, S.J. Lee, J.Y. Lee, S.W. Kim, Ch.B. Lee, W.Y. Lee, M.S. Yoon, Int. J. Antimicrob. Agents 19, 576 (2002)
S.C. Cunnane, Zinc: Clinical and Biochemical Significance (CRC Press, Boca Raton, 1988)
C.L. Feucht, B.S. Allen, D.K. Chalker, J. Am. Acad. Dermatol. 3, 483 (1980)
H. Icbudak, Z. Heren, D.A. Köse, H. Necefoğlu, J. Therm. Anal. Cal. 76, 837 (2004)
D.A. Köse, H. Necefoğlu, J. Therm. Anal. Cal. 93(2), 509 (2008)
D.A. Köse, A. Kaya, H. Necefoğlu, Russ. J. Coord. Chem. 33(6), 422 (2007)
D.A. Köse, H. İçbudak, H. Necefoğlu, Hacettepe J. Biol. Chem. 35(2), 123 (2007)
T. Hökelek, H. Necefoğlu, Acta Cryst. E58, m121 (2002)
W. Wolodkiewicz, W. Bryska, Polish J. Chem. 71(1–3), 16 (1997)
V. Zeleñák, Z. Vargová, K. Györyová, Spectrochimica Acta A66, 262 (2007)
A. Erxleben, Coord. Chem. Rev. 246, 203 (2003)
H. Necefoglu, W. Clegg, A.J. Scott, Acta Cryst. E57, m465 (2001)
K. Nakamoto, Infrared and Raman Spectra of Inorganic and Coordination Compounds (Wiley and Sons, New York, 1978)
D.A. Köse, H. Necefoğlu, H. Icbudak, J. Coord. Chem. 61(21), 3508 (2008)
D.A. Köse, B. Zümreoglu-Karan, O. Şahin, O. Büyükgüngör, J. Mol. Struct. 789, 147 (2006)
V. Zeleñák, M. Sabo, W. Massa, P. Lewellyn, Inorg. Chim. Acta 357, 2049 (2004)
D.A. Köse, G. Gökçe, S. Gökçe, İ. Uzun, J. Therm. Anal. Cal. 95(1), 247 (2009)
G.M. Sheldrick, Acta Cryst. A64, 112 (2008)
L.J. Farrugia, J. Appl. Cryst. 32, 837 (1999)
A.L. Spek, J. Appl. Cryst. 36, 7 (2003)
J. Bernstein, R.E. Davis, L. Shimoni, N.L. Chang, Angew. Chem. Int. Ed. Engl. 34, 1555 (1995)
Acknowledgments
The authors wish to acknowledge the Faculty of Arts and Sciences, Ondokuz Mayıs University, Turkey, for the use of the Stoe IPDS-II diffractometer (purchased from Grant No. F279 of the University Research Fund).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Köse, D.A., Ay, A.N., Şahin, O. et al. A mononuclear, mixed (salicylato) (nicotinamide) complex of Zn(II) with penta- and hexa-coordination sites: a novel framework structure. J IRAN CHEM SOC 9, 591–597 (2012). https://doi.org/10.1007/s13738-012-0072-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13738-012-0072-9