Abstract
Context
Fir species are mainly distributed across the high latitudes, preferring cold climates. The phylogeographic origins of those subtropical fir species that occur in low latitudes in southern China remain elusive, as does the nature of any inter-lineage hybridization.
Aims
In fir species, mitochondrial DNA (mtDNA) is maternally inherited and chloroplast DNA (cpDNA) is paternally inherited, and the genetic variations in the two make them particularly useful for examining species’ hybridizations and evolutionary histories.
Materials and methods
We sequenced four DNA fragments: two of mtDNA and two of cpDNA for 161 individuals from four subtropical endangered fir species (Abies beshanzuensis, Abies ziyuanensis, Abies yuanbaoshanensis, and Abies fanjingshanensis) and two more northerly distributed ones (Abies recurvata and Abies fargesii) from central and southwest China.
Results
Three mitotypes and four chlorotypes were recovered from the four southern species. In most populations, individuals share a single fixed mitotype and chlorotype. Three mitotypes clustered into two distinct clades, each associated with one of the northern species examined. For four chlorotypes, one occurred in A. ziyuanensis, A. beshanzuensis, A. fargesii, and A. recurvata, another in A. ziyuanensis and A. recurvata, the remaining two differed from others by two mutations exclusively in A. fanjingshanenis and A. yuanbaoshanensis.
Conclusion
Phylogeographic origins of the subtropical fir species are complex, and genetic admixtures occurred during the evolutionary history of A. ziyuanensis. The genotyped populations further provide basic frames for genetic delimitation and effective conservation of these endangered species in the future.
Similar content being viewed by others
1 Introduction
Past climatic oscillations greatly affected the distributions of most plants (Hewitt 2004), although the effects varied between localities and groups of species (Avise 2004). Temperate plants generally retreated to refugia at low altitudes or latitudes when the climate became cold and recolonized their former ranges as temperatures subsequently rose (Avise 2004). However, such range shifts may still have left populations in nunatak refugia (glacial refugia on mountain peaks that protruded above the glaciers in the core of a mountain range) at high altitudes or in high latitudes during cold periods, as well as in isolated mountains at low latitudes when the climate warmed once more (Holderegger and Thiel-Egenter 2009; Stehlik et al. 2002). Any such isolated populations probably had extremely low genetic diversity and high genetic differentiation, and some may have finally evolved into new (cryptic or morphologically distinct) species due to the strong genetic drift caused by bottlenecks or founder effects (Avise 2004). In particular, lineages that had retreated or recolonized from different areas or refugia usually hybridized with one another as their ranges merged, producing numerous hybrid populations or species (e.g., Abbott et al. 2010).
Fir species usually occur in the Northern Hemisphere in high latitude regions or at high altitudes. The forest ecosystems dominated by fir trees provide a basic home for a great diversity of animals and plants (Farjon and Rushforth 1989; Florin 1963; Liu 1971). However, a limited number of fir species are found further south in subtropical mountains; such species occur in discrete, disconnected populations (Florin 1963; Liu 1971). In China, there are 22 fir species, most of which are found in high latitude or high altitude regions in northern, central, or southwest China (Fu et al. 1999). However, in southern China, there are four highly endangered species: Abies beshanzuensis M. H. Wu, Abies ziyuanensis L. K. Fu & S. L. Mo, Abies yuanbaoshanensis Y. J. Lu & L. K. Fu, and Abies fanjingshanensis W. L. Huang (Fu et al. 1999) (Fig. 1). These species have a very restricted distribution on the subtropical mountain tops, and the very small numbers of surviving individuals mean that they are extremely endangered (Farjon 1990; Liu 1971; Ning et al. 2005; Xiang 2001; Zhang et al. 2004). These four species have been classified under two infrageneric sections: A. beshanzuensis and A. ziyuanensis were placed in Sect. Momi, which is morphological similarity to the Abies recurvata complex that occurs in southwest China; Abies fanjingshaensis and Abies yuanbaoshanensis were placed in Sect. Pseudopicea, which is similar to the Abies fargesii complex from central China (Farjon and Rushforth 1989). Phylogenetic analyses based on internal transcribed spacer data suggested that these two sections clustered into monophyletic clades but with low bootstrap support (Xiang et al. 2009). It has often been suggested that these subtropical fir species originated from southward retreats of the fir forests of central or southwest China (Florin 1963; Li 1995; Liu 1971; Liu et al. 2002; Ying 1989). However, actual phylogeographic relationships between them and those species occurring further north remain unknown. Did they originate only from the fir forests of central China or from southwest China or from both areas? If the southward retreats of fir forests from two regions created these subtropical species, did introgressions or hybridization occur between them?
In order to answer these questions, we sequenced mitochondrial (mt) and chloroplast (cp) DNA fragments from four subtropical and two northern species from different regions. Like most other conifers, these cytoplasmic types of DNA in Abies are haploid; mtDNA is maternally transmitted via seed while cpDNA is paternally transmitted via pollen (Liepelt et al. 2002). Thus, these DNA data together provide a sound basis for tracing range shifts (via seed) during retreats and recolonizations and for identifying potential hybrid origins of the current species or populations through maternal and paternal relationships (e.g., Liepelt et al. 2002, 2010; Song et al. 2003).
2 Materials and methods
2.1 Sampling design
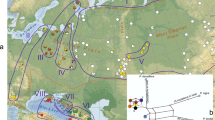
Samples were obtained from 161 individuals of 19 populations spanning the geographic range of each of the six focal species (see Fig. 1 and Table S1). Of these samples, 38 individuals (19.3% in total) of six populations representing four southern species were sampled due to their limited distribution. For example, there are only three surviving individuals of A. beshanzuensis (Ning et al. 2005; Zhang et al. 2004); we sampled two adult individuals. More populations and individuals of A. ziyuanensis are known. We found and collected all three mature individuals at Yinzhulaoshan. The other two known populations contain more adult individuals (more than 20). In each population, therefore, we sampled seven mature individuals that were growing at least 20 m apart; the other individuals were growing very close to the sampled trees. Only one population of A. yuanbaoshanensis was found (Ning et al. 2005; Zhang et al. 2004), and we located fewer than 20 adult individuals; we collected material from seven individuals each about 10 m from the next. The only known population of A. fanjingshanensis is large and contains more than 50 mature individuals; we therefore sampled 12 adult individuals that were about 100 m apart. For the two more widely distributed, northerly species, we randomly chose six or seven populations from across their entire range. In principal, 10 individuals separated by at least 100 m were collected from each population. A palm Extrex GIS (Garmin) was used to measure the latitude, longitude, and altitude of each sampling site.
2.2 DNA extraction, amplification, and sequencing
Total genomic DNA was extracted from a sample of approximately 20 mg of silica gel-dried needles, using the hexadecetytrmethyl ammonium bromide procedure (Doyle and Doyle 1987). Intron 4 of subunit 5 of the mitochondrial nicotinamide adenine dinucleotide dehydrogenase gene (nad5 intron 4) and Intron 1 of subunit 7 of the same gene (nad7 intron 1) were amplified and sequenced, using primers described by Wu et al. (1998) and Jaramillo-Correa et al. (2004), respectively. In addition, cpDNA trnS-G and trnL-F were amplified and sequenced following Demesure et al. (1995) and Taberlet et al. (1991), respectively. Polymerase chain reactions (PCRs) were performed in a 25-μL volume, which contained 10–40 ng plant DNA, 50 mM Tris–HCl, 1.5 mM MgCl2, 250 μg/mL BSA, 0.5 mM dNTPs, 2 μM of each primer, and 0.75 U Taq polymerase. The PCR amplifications were conducted using the following temperature profile: 4 min at 94°C, followed by 37 cycles of 40 s at 94°C, 40 s of annealing at 58°C (for mtDNA) or 60°C (for cpDNA), and 1.2 min at 72°C, with a final 7 min extension at 72°C.
We then purified the PCR products using a TIANquick Midi Purification Kit (TIANGEN, Beijing, China). Sequencing reactions were performed with the PCR primers described above using an ABI PRISM BigDye Terminator version 3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). We aligned the sequences obtained with Clustal X (Thompson et al. 1997), following manual modifications in MEGA4 (Tamura et al. 2007). The newly recovered sequences have been deposited in the NCBI Genbank under Accession Numbers JN584218-JN584263. In addition, the accession numbers of A. fargesii sequences have been supported in a previous study (see Wang et al. 2011).
2.3 Phylogenetic and phylogeographic analyses
The limited numbers of mutations associated with mtDNA and cpDNA in firs preclude the construction of phylogenetic trees using the maximum parsimony and maximum likelihood methods. We therefore followed previous researchers (Liepelt et al. 2002, 2010; Jaramillo-Correa et al. 2004, 2008) and constructed minimum-spanning trees for the recovered haplotypes using the software Network version 4.2.0.1 (available at http://www.fluxus–engineering.com; Bandelt et al. 1999). For phylogeographic analyses, we defined three regions based on species distributions: southwest China (SWC), central China (CTC), and southern China (STC) (See Fig. 1 and Table S1). Population structure within and between regions was analyzed separately for mtDNA and cpDNA. The average gene diversity within populations (H S), total gene diversity (H T), and the coefficients of differentiation G ST and N ST were estimated for each species. G ST is calculated solely on the basis of the frequencies of haplotypes, whereas N ST takes into account the similarities (phylogenetic relatedness) between haplotypes; calculations were performed using PERMUT (Pons and Petit 1996; available at http://pierroton.inra.fr/labo/software/permutcpssr). Hierarchical partitioning of diversity among species, populations, and individuals was based on analyses of molecular variance (AMOVA), using the software Arlequin version 3.1 (Excoffier et al. 2006), with significance tests based on 1,000 permutations.
3 Results
3.1 mtDNA variation and distribution
Polymorphisms were recorded within both mtDNA regions examined. Three variants of nad7 intron 1 were the result of one indel, and four variants of nad5 intron 4 were recovered as a result of one indel and three nucleotide substitutions (Table S2). By considering the two polymorphic mtDNA regions together, we identified five distinct haplotypes (I–V), clustering into two distinct clades in the minimum-spanning tree, separated by five mutations (Fig. 1). Mitotype I, which represented one distinct clade, was found widely across four species: A. yuanbaoshanensis, A. fanjingshanensis, A. ziyuanensis, and A. recurvata. However, four remaining mitotypes belonging to the other clade were found in A. beshanzuensis (IV), A. ziyuanensis (V), and A. fargesii (II and III), respectively (Fig. 1).
Low levels of genetic variation were detected within the fir species studied herein using two mitochondrial fragments. No polymorphism was identified within A. recurvata, A. yuanbaoshanensis, A. fanjingshanensis, or A. beshanzuensis (Fig. 1 and Table S1). However, all populations of A. ziyuanensis are monomorphic with two different mitotypes fixed.
3.2 cpDNA variation and distribution
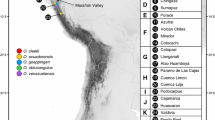
One indel and 13 substitutions were found in two cpDNA sequences, which combined into 17 chlorotypes (A–Q) (Fig. 2 and Table S2). These chlorotypes can be tentatively divided into three clades. The first comprises those chlorotypes recovered from A. fargesii (A–H), one of which (A) was also present in the southern species. The second includes those chlorotypes from A. recurvata, one of which (I) was also present in one southern species. The third clade comprises the two chlorotypes (L and M) that are found exclusively in the southern species. This clade obviously originated from the second clade belonging to A. recurvata because L is closely related to I, being separated by only one step (Fig. 2). The two widely distributed chlorotypes in the former two clades (A and I) are separated by only one step. Only four of 17 chlorotypes were found in the four southern species. Chlorotype A was present in A. beshanzuensis and two populations of A. ziyuanensis (Fig. 2). This chlorotype is closely related to others occurring exclusively in A. fargesii (for example, chlorotype B). Another chlorotype I was found to have a discontinuous geographical distribution; it was present in one population of A. ziyuanensis as well as in A. recurvata in southwest China. The final two chlorotypes (L and M) found in the southern China were restricted to A. yuanbaoshanensis and A. fanjingshanensis. A total of 14 chlorotypes were found in the two widespread northern species, A. fargesii and A. recurvata.
The two widespread northern species (A. fargesii and A. recurvata) exhibit high diversity and have numerous private chlorotypes, while all the southern species exhibit extremely low polymorphism (Table S1). In addition, as with the mtDNA, genetic variation within both populations and species is extremely low. However, genetic variation between the populations of A. ziyuanensis is high because each is associated with a different chlorotype (Fig. 2 and Table 1).
4 Discussion
Our population genetic analyses of the four fir species in southern China based on mtDNA and cpDNA variations suggested that these species have complex phylogeographic origins. Especially, we found distinct genetic admixtures possibly due to hybridization and introgression in one of them (A. ziyuanensis). In addition, range expansions based on the genetic data are distinct in two northern species, indicating that they might have retreated southward and recolonized northward in response to the past climatic changes. The southward retreats of the northerly distributed species probably gave rise to the production of these subtropical fir species. The genotyped population data further provide basic frames for genetic delimitation and effective conservation of these endangered subtropical fir species in the future.
4.1 Phylogeographic history
Our results suggested that two major mitotypes, one from each of two clades, characterized the two widespread and northerly fir species (Fig. 1). However, the mitotypes recovered from A. beshanzuensis (IV) and one population of A. ziyuanensis (V) belonged to the same clade as those (II and III) recovered from A. fargesii while A. yuanbaoshanensis, A. fanjingshanensis, and two populations of A. ziyuanensis shared a mitotype (I) with the northern species A. recurvata. Given that the two clades differed by four mutations, the fragmentation of the ancestral genetic pool cannot have occurred very recently although we could not exclude it totally. If we assume that these two clades formed before the colonization of southern China (Florin 1963; Li 1995; Liu 1971; Liu et al. 2002; Ying 1989), the phylogenetic relationships between mitotypes indicate that the fir species in southern China possibly originated from two different more northerly regions, namely central and southwest China. In fact, the chlorotype networks seem to support this possibility. Chlorotype A is associated with two species, A. beshanzuensis and A. ziyuanensis, and it is also found widely in A. fargesii in central China. Similarly, chlorotype I was found in A. ziyuanensis and A. recurvata, which is present in southwest China. However, two chlorotypes found in A. yuanbaoshanensis and A. fanjingshanensis did not appear in the two northerly species. These two chlorotypes comprised a separate clade in the network, and they are most closely related to chlorotype I. These two chlorotypes probably originated independently in southern China from an ancestral chlorotype resembling I. In addition, it should be noted that more haplotypes at both mtDNA and cpDNA might have existed but vanished with the range shrinkage of these southern fir species. Therefore, our analyses suggested these southern fir species have complex phylogeographic origins, probably from southern retreats of the northern species, occurring in CTC, SWC, or other northerly distributed regions.
For the two northerly species, our population genetic analyses of both mtDNA and cpDNA data suggested that they had experienced extensive range expansions in the recent past. All populations of each species were dominated by a single mitotype (Fig. 1). This is consistent with numerous reports that range expansion from local or remote refugia promoted the widespread fixing of a single haplotype for mtDNA in conifers, which is associated with few mutations (Avise 2004; Du et al. 2009). However, a range expansion could have also resulted in a dominant haplotype with multiple rare alleles (a star-phylogeny pattern) for DNA fragments with a fast mutation rate (for example, cpDNA in conifers) (Avise 2004). In both A. recurvata and A. fargesii, we found numerous rare chlorotypes, possibly supporting an expansion hypothesis, although there was no distinct star-phylogeny pattern. The actual mutation rates of mtDNA and cpDNA in the Abies genus are not known. It is also not yet known when the fir species from the central and southwest China colonized southwards as a result of decreasing temperature and when subsequent expansions occurred as the climate warmed. However, all these genetic data seem to be consistent with previous speculations that the southward retreats of the northerly distributed species during the cold climate age gave rise to the production of the subtropical fir species in southern China (Florin 1963; Li 1995; Liu 1971; Liu et al. 2002; Ying 1989).
4.2 Genetic delimitation and hybridization
Our genetic analyses suggested that a combination of the recovered mitotypes and chlorotypes could be used to characterize three of the four rare species that we examined; the exception was A. ziyuanensis. Mitotype IV was only found in A. beshanzuensis, although this species shared chlorotype A with A. ziyuanensis and A. fargesii. Although A. yuanbaoshanensis and A. fanjingshanensis shared a mitotype (I) with A. ziyuanensis and A. recurvata, chlorotype L was fixed in both of them, while chlorotype M was only found in A. fanjingshanensis. Although these two species can be distinguished from the other fir species and from each other by the presence of a single species-specific chlorotype (M) (Du et al. 2009; Tsumura and Suyama 1998), taxonomic and genetic delimitations between them need further studies based on more data.
As noted before, the networks of both mitotypes and chlorotypes suggested that these three species probably originated from different southward movements of fir species: A. beshanzuensis from the A. fargesii complex-like ancestors in central China and A. yuanbaoshanensis–A. fanjingshanensis from the A. recurvata complex-like ancestors in southwest China. However, three populations of A. ziyuanensis contained both mitotypes and chlorotypes originating from both regions. For example, two populations (15 and 17) were characterized by mitotype I in the clade with A. recurvata from southwest China, while the third population (16) was characterized by mitotype V, which is in the other clade containing A. fargesii from central China. Meanwhile, two populations (16 and 17) shared the same chlorotype (A) with A. fargesii while the other population (15) had chlorotype I, as did A. recurvata. It is obvious that both the mitotype and chlorotype of the eastern population (16) had common origins with A. beshanzuensis and A. fargesii from central China, while the western population (15) shared common mitotype and chlorotype origins with A. yuanbaoshanensis, A. fanjingshanensis, and A. recurvata from southwest China. However, the central population (17) contained two ancestral lineages based on its different maternal (mitotype) and paternal (chlorotype) origins. Because all populations and individuals of A. ziyuanensis are morphologically uniform and distinct from other species (Farjon and Rushforth 1989; Fu et al. 1999; Liu 1971), two possible scenarios may account for the genetic pattern identified for this species. First, A. ziyuanensis may be a paraphyletic complex composed by two different lineages. It is evolutionary convergence due to the local environmental constraints that resulted in morphological similarity between different lineages. Second, it is more likely that hybridization or introgressions caused such genetic pattern. For example, this species originated from diploid hybridizations between two different ancestral lineages. Because of the different backcross frequencies, each population may have randomly fixed mitotypes or chlorotypes from paternal or maternal lineages. It is very common in homoploid hybrid species that different populations fixed different chlorotypes or mitotypes from both parental lineages (e.g., Rieseberg et al. 2007; Song et al. 2003). Alternatively, this species originated from one of two ancestral lineages from further north. However, after its establishment, mitotypes or chlorotypes from the other lineage might have introgressed totally or partially into a few populations of A. ziyuanensis. These analyses suggested that hybridizations or introgressions between two ancestral lineages that colonized southern China might have occurred during the evolutionary history of A. ziyuanensis in response to climatic oscillations.
Our genetic characterization and the inferred interspecific relationships are largely inconsistent with previous studies based on morphological traits (Farjon and Rushforth 1989). Morphologically, A. fanjingshaensis and A. yuanbaoshanensis are similar and seem to be closely related to A. fargesii (all within Sect. Pseudopicea) from central China. In addition, A. beshanzuensis, A. ziyuanensis, and A. recurvata are rather similar to one another, together belonging to the same section (Sect. Momi). This inconsistency between the morphological and the genetic evidence may suggest that convergent evolution in morphology may have occurred within these species. However, further studies are needed to clarify the underlying mechanisms.
4.3 Intraspecific diversity and conservation
In general, the levels of genetic diversity for mtDNA and cpDNA within the four southern Abies species examined (A. beshanzuensis, A. ziyuanensis, A. yuanbaoshanensis, and A. fanjingshanensis) were similar (Table S1) but obviously lower than those of other conifers (e.g., Du et al. 2009; Jaramillo-Correa et al. 2008; Tsumura and Suyama 1998). In addition, AMOVA indicated that most of the total variation in mtDNA and cpDNA makers occurred among species or among populations of each species (Table 1). The low genetic diversity within populations and high genetic differentiation between populations (Table 2) are also consistent with a recent study of A. ziyuanensis based on intersimple sequence repeat and amplified fragment length polymorphism markers (Tang et al. 2008). This could result mainly from strong genetic drift associated with the presence of small isolated populations during past climatic oscillations after the establishment of these species (Avise 2004). The four southern species examined have been classified as being endangered (Frankham et al. 2002). Unfortunately, habitats suitable for these species continue to deteriorate (Xiang 2001). The total number of surviving individuals for each population/species is still decreasing according to our field investigations. Because of the extremely endangered status of each species, every individual (including those rare seedlings) from any population should be rigorously protected.
References
Abbott RJ, Brennan AC, Hegarty MJ, Hiscock SJ (2010) Homoploid hybrid speciation in action. Taxon 59:1375–1386
Avise JC (2004) Molecular markers, natural history, and evolution. Sinauer, Sunderland
Bandelt HJ, Forster P, Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Demesure B, Sodzi N, Petit RJ (1995) A set of universal primers for amplification of polymorphic non-coding regions of mitochondrial and chloroplast DNA in plants. Mol Ecol 4:129–131
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Du FK, Petit RJ, Liu JQ (2009) More introgression with less gene flow: chloroplast vs mitochondrial DNA in the Picea asperata complex in China, and comparison with other Conifers. Mol Ecol 18:1396–1407
Excoffier L, Laval G, Schneider S (2006) ARLEQUIN ver 31: an integrated software package for population genetics data analysis. Computational and Molecular Population Genetics Lab (CMPG), Institute of Zoology, University of Berne, Berne
Farjon A (1990) Pinaceae. Koeltz Scientific Book, Germany
Farjon A, Rushforth KD (1989) A classification of Abies Miller (Pinaceae). Notes Roy Bot Gard Edinburgh 46:59–77
Florin R (1963) The distribution of conifer and taxad genera in time and space. Acta Hort Berg 20:194–256
Frankham R, Ballou JD, Briscoe DA (2002) Introduction to Conservation Genetics. Cambridge University Press, Cambridge
Fu LK, Li N and Robert RM (1999) Sections on Cephalotaxaceae, Ginkgoaceae and Pinaceae. In: Wu ZY and Peter HR (ed) Flora of China, Volume 4. Science Press, Beijing
Hewitt GM (2004) Genetic consequences of climatic oscillations in the Quaternary. Phil Trans R Soc Lond B 359:183–195
Holderegger R, Thiel-Egenter C (2009) A discussion of different types of glacial refugia used in mountain biogeography and phylogeography. J Biogeogra 36:476–480
Jaramillo-Correa JP, Beaulieu J, Bousquet J (2004) Variation in mitochondrial DNA reveals mutilple distant glacial refugia in black spruce (Picea mariana), a transcontinental North American conifer. Mol Ecol 13:2735–2747
Jaramillo-Correa JP, Aguirre-Planter E, Khasa DP, Eguiarte LE, Pinero D, Furnier GR, Bousquet J (2008) Ancestry and divergence of subtropical montane forest isolates: molecular biogeography of the genus Abies (Pinaceae) in southern Mexico and Guatemala. Mol Ecol 17:2476–2490
Li N (1995) Study on the geographic distribution, origin and dispersal of the family Pinaceae. Lindl Acta Phytotaxon Sin 33:105–130
Liepelt S, Bialozyt R, Ziegenhagen B (2002) Wind-dispersed pollen mediates postglacial gene flow among refugia. Proc Natl Acad Sci U S A 99:14590–14594
Liepelt S, Mayland-Quellhorst E, Lahme M, Ziegenhagen B (2010) Contrasting geographical patterns of ancient and modern genetic lineages in Mediterranean Abies species. Plant Syst Evol 284:141–151
Liu TS (1971) A monograph of the genus Abies. The Department of Forestry College of Agriculture, National Taiwan University, Taipei
Liu ZL, Fang JY, Piao SL (2002) Geographical distribution of species in genera Abies, Picea and Larix in China. Acta Geogr Sin 57:577–586
Ning S, Tang R, Cao J (2005) Current status and conservation countermeasures of germplasm resources of Abies ziyuanensis. Guihaia 25:191–200 (in Chinese with English abstract)
Pons O, Petit RJ (1996) Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144:1237–1245
Rieseberg LH, Kim SC, Randell RA, Whitney KD, Gross BL, Lexer C, Clay K (2007) Hybridization and the colonization of novel habitats by annual sunflowers. Genetica 129:149–165
Song BH, Wang XQ, Wang XR, Ding KY, Hong DY (2003) Cytoplasmic composition in Pinus densata and population establishment of the diploid hybrid pine. Mol Ecol 12:2995–3001
Stehlik I, Blattner FR, Hokderegger R, Bachmann K (2002) Nunatak survival of the high Alpine plant Eritrichium nanum (L.) Gaudin in the central Alps during the ice ages. Mol Ecol 11:2027–2036
Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol 17:1105–1109
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 40. Mol Biol Evol 24:1596–1599
Tang S, Dai W, Li M, Zhang Y, Geng Y, Wang L, Zhong Y (2008) Genetic diversity of relictual and endangered plant Abies ziyuanensis (Pinaceae) revealed by AFLP and SSR markers. Genetica 133:21–30
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acid Res 25:4876–4882
Tsumura Y, Suyama Y (1998) Differentiation of mitochondrial DNA polymophisms in populations of five Japanese Abies species. Evolution 52:1031–1042
Wang J, Abbott RJ, Peng YL, Du FK, Liu JQ (2011) Species delimitation and biogeography of two fir species (Abies) in central China: cytoplasmic DNA variation. Heredity 107(4):362–370. doi:10.1038/hdy.2011.22
Wu JY, Krutovskii KV, Strauss SH (1998) Abundant mitochondral genomediversity, population differentiation and convergent evolution in pines. Genetics 150:1605–1614
Xiang QP (2001) A preliminary survey on the distribution of rare and endangered plants of Abies in China. Guihaia 21:113–117 (in Chinese with English abstract)
Xiang QP, Xiang QY, Guo YY, Zhang XC (2009) Phylogeny of Abies (Pinaceae) inferred from nrITS sequence data. Taxon 58:141–152
Ying TS (1989) Areography of the gymnosperms of China (1): distribution of the Pinaceae of China. Acta Phytotaxo Sin 27:27–38
Zhang YR, Luo JC, Gui XJ (2004) Study on the protection and breeding of the imminent danger species Abies ziyuanensis L. K. Fu. Huna Forest Sci Tech 31:26–29
Acknowledgements
We are grateful to two anonymous reviewers for their constructive suggestions. We thank John Blackwell for polishing the English of the final version.
Funding
This research was supported by grants from the Natural Science Foundation of China (Grant numbers 30725004) to J. Q. L.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: Christophe Plomion
Contribution of the co-authors
J.L. designed research; J.L., B.T., and G.R. collected samples; Y.P., J.W., Q.G, and B.T. performed research; Y.P. and J.L. analyzed data; and Y.P., S.Y. and J.L. wrote the paper.
Yanling Peng and Shuming Yin contributed equally to the work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
Locations of populations and sample sizes (N) of six closely related Abies spp. examined to determine mtDNA and cpDNA haplotypes. For each population, estimates of genetic diversity (H E) and nucleotide diversity (π) are given as a percentage (DOCX 20 kb)
Table S2
Variable sites of aligned sequences of two mtDNA fragments (nad7-1 and nad5-4) and two cpDNA fragments (trnS-G and trnL-F) from which five mitotypes and 17 chlorotypes were recorded across six closely related Abies spp. Sequences are numbered from the 5′ to the 3′ end in each region (DOCX 26 kb)
Rights and permissions
About this article
Cite this article
Peng, Y., Yin, S., Wang, J. et al. Phylogeographic analysis of the fir species in southern China suggests complex origin and genetic admixture. Annals of Forest Science 69, 409–416 (2012). https://doi.org/10.1007/s13595-011-0170-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13595-011-0170-3